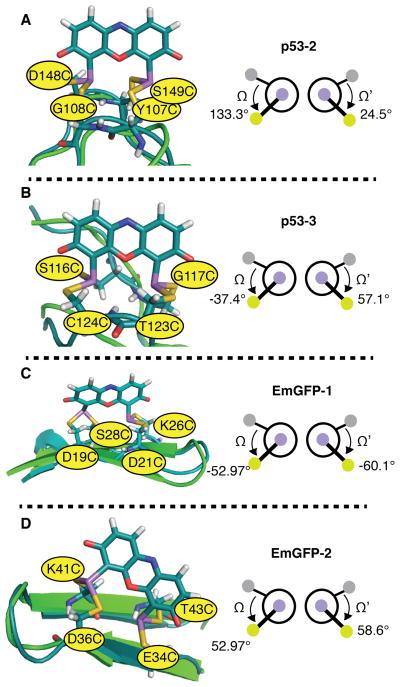
Figure 8.
Models of ReAsH in complex with previously reported Cys4 motifs within p53 and EmGFP and the associated values of Ω and Ω’: (A) p53-2; (B) p53-3; (C) EmGFP-1 and (D) EmGFP-2. Three of these proteins (p53-2, p53-3, and EmGFP-1) formed fluorescent complexes with ReAsH, while one (EmGFP-2) did not. Hydrogen bonding networks are not shown for clarity. In each case, the minimized structure of the indicated protein variant bound to ReAsH is shown in teal and aligned with the native structure shown in green. As discussed in the text, although the Ω and Ω’ angles for the minimized EmGFP-2 structure fall within the fluorescent range, EmGFP-2 binds ReAsH poorly and the minimized structure contains a disrupted β-strand network.