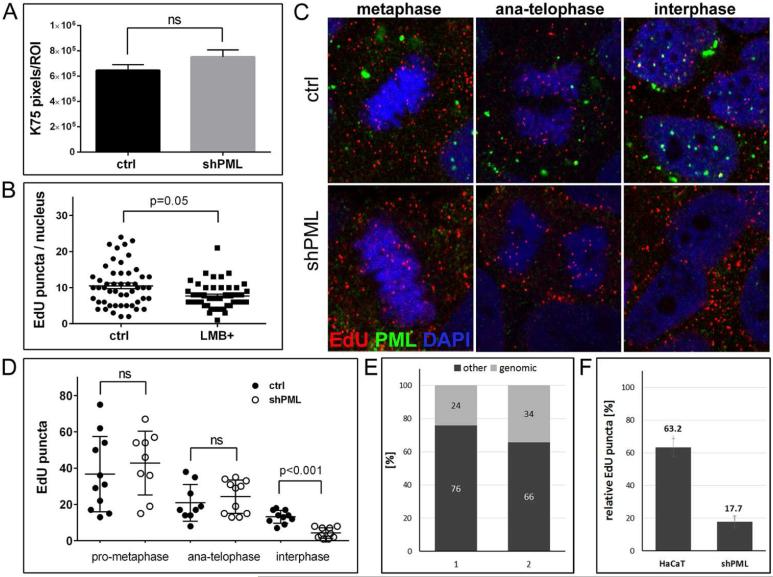
Fig 3. Binding, uncoating, and delivery of HPV16 pseudogenome to the nucleus of PML-deficient HaCaT cells.
(A) Cell-binding assay. HPV16 pseudoviruses were bound to of HaCaT cells stably transduced with lentiviruses expressing scrambled or shRNA directed against PML for 1 h and L1 was detected using K-75 antibody. (B) Quantification of EdU-labeled viral pseudogenome in the nuclei of PML-deficient HaCaT cells after inhibition of nuclear export. Cells were grown on glass coverslips and infected with EdU-labeled PsV and processed at 26 hpi. Leptomycin B (10ng/ml) was added 2 h prior to fixation. Cells were stained for EdU, PML, lamin A/C and DAPI. Number of EdU-labeled viral pseudogenome was quantified in z-stacks spanning the whole nucleus in 51 control and 47 LMB-treated cells. Student's t-test was used to determine statistical significance of the data. (C) EdU-labeled viral pseudogenome in different stages of mitosis. Cells were grown on glass coverslips and infected with EdU-labeled PsV and processed at 24 hpi for the detection of EdU-labeled DNA (red), PML (green), lamin A/C (not shown) and DAPI (blue). A single slice from a z stack is shown. (D) Number of EdU-labeled viral genome bound to cellular chromatin or in the nucleus was scored in cells captured in different stages of mitosis. An example representative of two independent experiments is shown. Results were calculated from at least 30 cells per experiment. Student's t-test was used to determine statistical significance of the data. (E) Composition of encapsidated DNA in quasivirion preparations according to MiSeq Reporter. Two samples of DNA isolated from independent quasivirion preparations were sequenced using Illumina NextSeq 500. Results of alignment to the human genome are shown as percent of total sequenced encapsidated DNA. (F) Quantification of EdU-labeled viral genome bound to cellular chromatin in cells at interphase normalized to anaphasetelophase (shown in (D)).