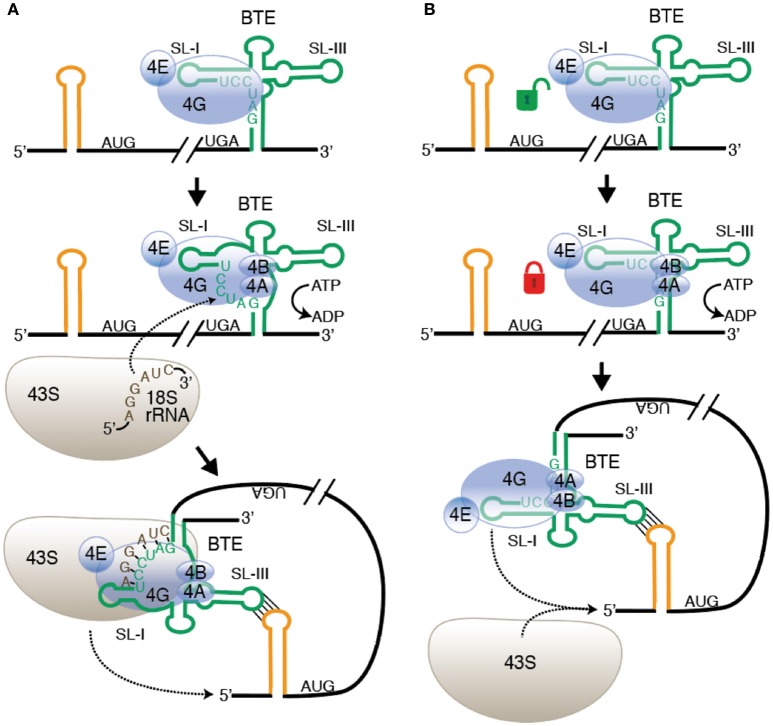
Figure 2.
Alternative models of ribosome recruitment and delivery to the 5′-UTR via the BTE. (A) Base pairing to rRNA model. Top: eIF4F binds to SL-I of the BTE (green) through the eIF4G subunit. eIF4E enhances but is not required for BTE binding. Middle: Helicase (eIF4A + eIF4B) binds and uses ATP hydrolysis to unwind GAUCCU, making it available to base pair to 18S rRNA at a conserved sequence in the region where the Shine-Dalgarno binding site is located in prokaryotic 16S rRNA. Bottom: The 43S preinitiation complex base pairs to the BTE and is delivered to the 5′ end by long-distance base pairing (yellow stem-loop). (B) Conventional ribosome recruitment model. Top: eIF4F binds BTE as in (A). Middle: Binding of eIF4A + eIF4B and ATP hydrolysis increases binding affinity of eIF4F, “locking” it on to the BTE, perhaps by altering the structure of BTE RNA. Bottom: eIF4 complex is delivered to 5′ end by long-distance base pairing where it recruits the 43S preinitiation complex to the RNA. In both models, 43S scanning from the 5′ end to the start codon is the same as in normal cap-dependent translation. Not shown: other factors, such as eIF3 and factors in the preinitiation complex.