Figure 3.
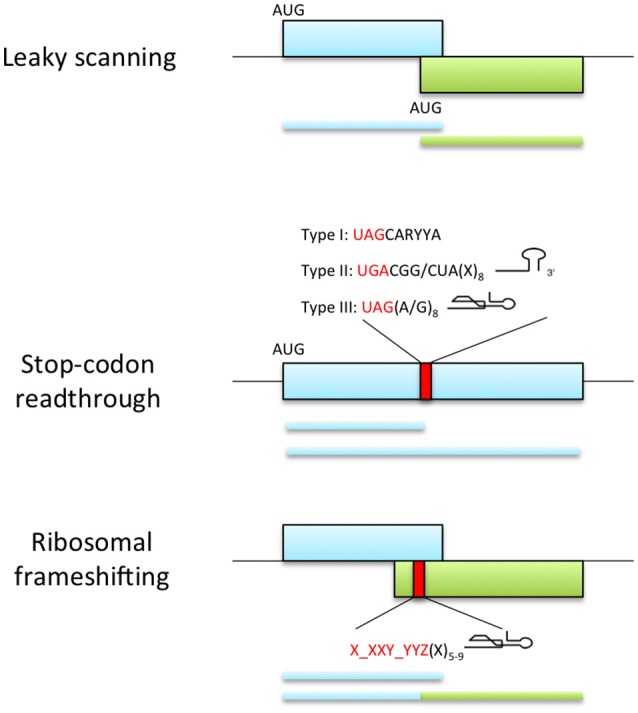
Viral recoding strategies. Top panel represents leaky scanning mechanism where ribosomes fail to start translation at the first AUG codon and continue scanning until they reach an alternative start codon in the optimal initiation context. This process allows the expression of two proteins with distinct amino acid sequence when the initiation sites are in different reading frames (as shown) or C-terminally coincident isoforms of a single protein if initiation sites are in-frame (not shown). Middle panel shows the expression of proteins with alternative C-terminal because a portion of ribosomes fail to terminate at a stop codon and continue translation. The efficiency of readthrough can be stimulated by the presence of elements downstream of the stop codon: UAG stop codon followed by the consensus motif CARYYA, where R is a purine and Y is a pyrimidine (Type I); UGA stop codon followed by CGG or CUA triplet and a stem-loop structure separated from the stop codon by 8 nt (Type II); UAG stop codon and adjacent G or purine octanucleotide and a compact pseudoknot structure (Type III). Bottom panel represents ribosomal frameshifting strategy, where ribosomes are directed into a different reading frame guided by the slippery signal X_XXY_YYZ (X and Y can be any base and Z is any base except G) and a secondary structure element located 5-9 nt downstream the slippery sequence.
