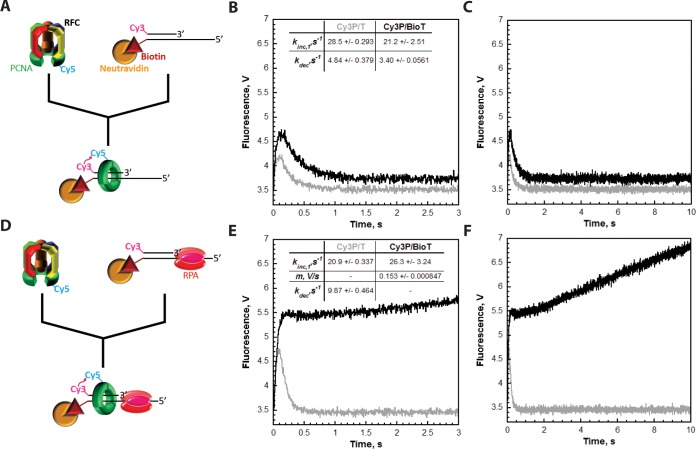
Figure 3.
RFC releases PCNA onto the duplex region of P/T DNA. Each FRET trace represents the average of at least eight shots. For comparison, all FRET traces are normalized to the same starting value (at t = 0.005 s). (A) Schematic representation of the experimental procedure for panel B. (B) Cy5-PCNA (110 nM homotrimer) was preincubated with RFC (110 nM) and ATP. This preformed RFC·Cy5-PCNA·ATP complex was mixed with Cy3-labeled DNA (100 nM Cy3P/BioT or Cy3P/T) and ATP in a stopped-flow instrument, and the FRET signal was followed. The loading traces for the Cy3P/T (gray) and Cy3P/BioT DNA (black) substrates are shown, and each was fit to a double-exponential equation. The calculated rate constants are reported for each. (C) Extended time courses (10 s) for each FRET trace from panel B. (D) Schematic representation of the experimental procedure for panel E. (E) The experiments depicted in panel B were repeated except the Cy3-labeled DNA was preincubated with excess RPA (242 nM). The loading trace for the Cy3P/T DNA substrate (gray) was fit to a double-exponential equation. The loading trace for the Cy3P/BioT DNA substrate (black) was fit to a single exponential and a linear phase. The kinetic values calculated from the fits for each trace are indicated. (F) Extended time courses (10 s) for each FRET trace from panel E.