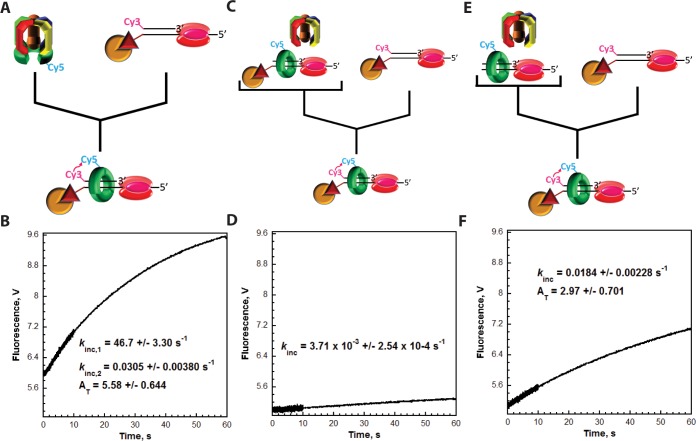
Figure 5.
Catalytic loading of PCNA is rate-limited by dissociation of RFC from RPA-coated ssDNA. Each FRET trace represents the average of at least eight shots. For comparison, all FRET traces are normalized to the same starting value (at t = 0.005 s). (A) Schematic representation of the experimental procedure for panel B. (B) Cy5-PCNA (180 nM homotrimer) was preincubated with RFC (180 nM) in the presence of ATP. This preformed RFC·Cy5-PCNA·ATP complex was mixed with Cy3P/BioT DNA (200 nM), RPA (285 nM), and ATP in a stopped-flow instrument, and the FRET signal was followed. The loading trace was fit to a double-exponential equation, and the kinetic values are reported. (C) Schematic representation of the experimental procedure for panel D. (D) Unlabeled P/BioT DNA (200 nM) was preincubated with RPA (285 nM), Cy5-PCNA (180 nM homotrimer), RFC (180 nM), and ATP. This solution was then mixed with Cy3P/BioT DNA (200 nM), RPA (285 nM), and ATP in a stopped-flow instrument, and the FRET signal was followed. The loading trace was fit to a single-exponential equation, and the rate constant is reported. (E) Schematic representation of the experimental procedure for panel F. (F) The experiments depicted in panels C and D were repeated except RPA (285 nM), Cy5-PCNA (180 nM homotrimer), RFC (180 nM), and ATP were preincubated with unlabeled P/T DNA (200 nM). The loading trace was fit to a single-exponential equation, and the kinetic values are reported.