In the title biphenyl derivative, the dihedral angle between the benzene rings is 52.84 (10)°. In the crystal, molecules are linked by two pairs of N—H⋯O hydrogen bonds, forming chains propagating along [101].
Keywords: crystal structure, dinitro, biphenyl, amine, biphenyl derivatives, hydrogen bonding
Abstract
In the title biphenyl derivative, C12H9N3O4, the dihedral angle between the benzene rings is 52.84 (10)°. The nitro group attached to the benzene ring is inclined to the ring by 4.03 (2)°, while the nitro group attached to the amino-substituted benzene ring is inclined to the ring by 8.84 (2)°. In the crystal, molecules are linked by two pairs of N—H⋯O hydrogen bonds, forming chains propagating along [101]. Within the chains, these N—H⋯O hydrogen bonds result in the formation of R 2 2(20) and R 2 2(14) ring motifs. The latter ring motif is reinforced by a pair of C—H⋯O hydrogen bonds, enclosing R 2 1(6) ring motifs. The chains are linked by a second C—H⋯O hydrogen bond, forming a three-dimensional supramolecular structure.
Chemical context
Biphenyl and its derivatives have been shown to play an important role in fighting cancer and arteriosclerosis in humans (Umeda et al., 2005 ▸). The dihedral angle between the phenyl rings of biphenyl derivatives is associated with their affinity for cellular target molecules and, therefore, can correlate with their toxicity. The parent compound, biphenyl, adopts a planar conformation in the solid state with a dihedral angle of 0° (Trotter, 1961 ▸). The calculated dihedral angle for biphenyl derivatives without ortho substituents is ca 41° (Shaikh et al., 2008 ▸). Deviations from the energetically most favourable conformation are most likely the result of crystal packing effects, which allow such compounds to adopt an energetically favorable conformation in the solid state by maximizing the lattice energy. Many research groups have calculated the inter-ring torsion angle of biphenyl in the solid state (Brock, 1980 ▸; Brock & Minton, 1989 ▸; Bastiansen & Samdal, 1985 ▸), and in the gas phase (Bastiansen & Traetteberg, 1962 ▸). We report here a detailed description of the molecular structure and supramolecular features of the title biphenyl derivative, 4,4′-dinitro-[1,1′-biphenyl]-2-amine, (I).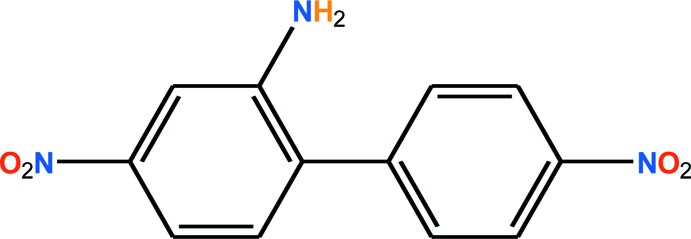
Structural commentary
The molecular structure of the title compound (I), is illustrated in Fig. 1 ▸. The dihedral angle between the two rings of the biphenyl unit is 52.84 (10)°. The nitro group (N3/O3/O4) is inclined to the benzene ring (C7–C12) to which it is attached by 4.03 (2)°. The nitro group (N1/O1/O2) is inclined to the amino-substituted benzene ring (C1–C6), to which it is attached, by 8.84 (2)°. The amino N atom, N2, lies in the plane of the C1–C6 benzene ring, and the N2—C5 bond length of 1.375 (3) Å clearly indicates a single bond. The C1—N1 distance of 1.466 (3) Å is slightly less than the C10—N3 bond distance of 1.477 (3) Å, which indicates that the 2-amino group containing a benzene ring (C1–C6) is more conjugated with the nitro group (N1/O1/O2) than is the other nitro group (N3/O3/O4) with respect to the C7–C12 benzene ring. The bond length of the C4—C7 bridge is 1.482 (3) Å, which indicates a single bond, and is similar to the same bond length of 1.494 (2) Å reported for dimethyl 2,2′-dinitrobiphenyl-4,4′-dicarboxylate (Lehane et al., 2014 ▸), and ca 1.493 Å observed in 2,2′-dinitrobiphenyl (Sekine et al., 1994 ▸).
Figure 1.
The molecular structure of the title compound, with the atom labelling. Displacement ellipsoids are drawn at the 40% probability level.
Supramolecular features
In the crystal, molecules are linked by two pairs of N—H⋯O hydrogen bonds, forming chains propagating along the [101] direction. Within the chains, these N—H⋯O hydrogen bonds result in the formation of  (20) and
(20) and  (14) ring motifs (Table 1 ▸ and Fig. 2 ▸). The latter ring motif is reinforced by a pair of C—H⋯O hydrogen bonds, enclosing
(14) ring motifs (Table 1 ▸ and Fig. 2 ▸). The latter ring motif is reinforced by a pair of C—H⋯O hydrogen bonds, enclosing  (6) ring motifs (Table 1 ▸ and Fig. 2 ▸). The chains are linked by a second C—H⋯O hydrogen bond (Table 1 ▸), forming a three-dimensional supramolecular structure, as illustrated in Figs. 3 ▸ and 4 ▸.
(6) ring motifs (Table 1 ▸ and Fig. 2 ▸). The chains are linked by a second C—H⋯O hydrogen bond (Table 1 ▸), forming a three-dimensional supramolecular structure, as illustrated in Figs. 3 ▸ and 4 ▸.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H2B⋯O1i | 0.92 (2) | 2.36 (2) | 3.229 (3) | 157 (2) |
| N2—H2A⋯O4ii | 0.89 (2) | 2.50 (2) | 3.345 (3) | 157 (2) |
| C6—H6⋯O1i | 0.93 | 2.54 | 3.308 (3) | 140 |
| C9—H9⋯O3iii | 0.93 | 2.57 | 3.496 (3) | 174 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Figure 2.
A view of the N—H⋯O and C—H⋯O hydrogen bonds (dashed lines; see Table 1 ▸), in the crystal of (I), forming chains that propagate along [101].
Figure 3.
A view along the b axis of the crystal packing of (I). Hydrogen bonds are shown as dashed lines (see Table 1 ▸) and, for clarity, only H atoms H2A, H2B, H6 and H9 have been included.
Figure 4.
A view along the a axis of the crystal packing of (I). Hydrogen bonds are shown as dashed lines (see Table 1 ▸) and, for clarity, only H atoms H2A, H2B, H6 and H9 have been included.
Database survey
A search of the Cambridge Structural Database (CSD, Version 5.38, update February 2017; Groom et al., 2016 ▸) revealed the structure of two similar compounds viz 4′-nitro-2-biphenylamine (II) (CSD refcode DIWFEU; Sutherland & Ali-Adib, 1986 ▸) and 4,4′-dinitrobiphenyl (III) (DNTDPH; Boonstra, 1963 ▸). In (II), the benzene rings are inclined to one another by 54.64 (6)°, compared to ca 32.91° in (III), and to 52.84 (2)° in the title compound (I). In (II), the nitro group is inclined to the benzene ring to which it is attached by 7.08 (6)°, compared to ca 3.55 and 10.14° in (III) and 8.3 (2)° in the title compound (I).
Synthesis and crystallization
The title compound (I), was prepared by a literature procedure (Ol’khovik et al., 2008 ▸). Orange prismatic crystals, suitable for single-crystal X-ray analysis, were grown by slow evaporation of a solution in ethanol.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. The N-bound H atoms were located in a difference Fourier map and refined with U iso(H) = 1.2U eq(N). The C-bound H atoms were included in calculated positions and refined as riding: C—H = 0.93–0.96 Å with U iso(H) = 1.2U eq(C).
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C12H9N3O4 |
| M r | 259.22 |
| Crystal system, space group | Monoclinic, P21/c |
| Temperature (K) | 296 |
| a, b, c (Å) | 14.2940 (11), 7.0352 (6), 11.6043 (9) |
| β (°) | 99.437 (6) |
| V (Å3) | 1151.15 (16) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.12 |
| Crystal size (mm) | 0.34 × 0.20 × 0.07 |
| Data collection | |
| Diffractometer | Stoe IPDS 2 |
| Absorption correction | Integration (X-RED32; Stoe & Cie, 2002 ▸) |
| T min, T max | 0.980, 0.993 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 6476, 2566, 1052 |
| R int | 0.044 |
| (sin θ/λ)max (Å−1) | 0.646 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.040, 0.092, 0.81 |
| No. of reflections | 2566 |
| No. of parameters | 180 |
| No. of restraints | 2 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.10, −0.12 |
Supplementary Material
Crystal structure: contains datablock(s) I, Global. DOI: 10.1107/S205698901700408X/su5355sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698901700408X/su5355Isup2.hkl
Supporting information file. DOI: 10.1107/S205698901700408X/su5355Isup3.cml
CCDC reference: 1537734
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors are grateful to the Ondokuz Mayıs University, Arts and Sciences Faculty, Department of Physics, 55139 Samsun, Turkey for X-ray the data collection.
supplementary crystallographic information
Crystal data
| C12H9N3O4 | F(000) = 536 |
| Mr = 259.22 | Dx = 1.496 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 14.2940 (11) Å | Cell parameters from 3570 reflections |
| b = 7.0352 (6) Å | θ = 2.1–27.8° |
| c = 11.6043 (9) Å | µ = 0.12 mm−1 |
| β = 99.437 (6)° | T = 296 K |
| V = 1151.15 (16) Å3 | Prism, orange |
| Z = 4 | 0.34 × 0.20 × 0.07 mm |
Data collection
| Stoe IPDS 2 diffractometer | 2566 independent reflections |
| Radiation source: sealed X-ray tube, 12 x 0.4 mm long-fine focus | 1052 reflections with I > 2σ(I) |
| Plane graphite monochromator | Rint = 0.044 |
| Detector resolution: 6.67 pixels mm-1 | θmax = 27.4°, θmin = 2.9° |
| rotation method scans | h = −18→18 |
| Absorption correction: integration (X-RED32; Stoe & Cie, 2002) | k = −8→9 |
| Tmin = 0.980, Tmax = 0.993 | l = −14→11 |
| 6476 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.040 | Hydrogen site location: mixed |
| wR(F2) = 0.092 | H atoms treated by a mixture of independent and constrained refinement |
| S = 0.81 | w = 1/[σ2(Fo2) + (0.0353P)2] where P = (Fo2 + 2Fc2)/3 |
| 2566 reflections | (Δ/σ)max < 0.001 |
| 180 parameters | Δρmax = 0.10 e Å−3 |
| 2 restraints | Δρmin = −0.12 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.61664 (11) | 0.3867 (2) | 0.93876 (16) | 0.0867 (5) | |
| N1 | 0.60769 (13) | 0.3912 (3) | 0.8317 (2) | 0.0701 (5) | |
| O3 | −0.08732 (12) | 0.2401 (3) | 0.36959 (17) | 0.1108 (7) | |
| O2 | 0.67457 (11) | 0.3941 (3) | 0.77943 (16) | 0.1004 (6) | |
| N2 | 0.26998 (16) | 0.4451 (3) | 0.8191 (2) | 0.0831 (6) | |
| N3 | −0.02768 (15) | 0.3445 (4) | 0.3418 (2) | 0.0899 (7) | |
| O4 | −0.04065 (12) | 0.4408 (3) | 0.25269 (19) | 0.1165 (7) | |
| C5 | 0.34478 (14) | 0.4181 (3) | 0.76003 (19) | 0.0575 (5) | |
| C6 | 0.43668 (14) | 0.4150 (3) | 0.82181 (19) | 0.0597 (5) | |
| H6 | 0.447162 | 0.429120 | 0.902589 | 0.072* | |
| C4 | 0.33093 (14) | 0.3924 (3) | 0.63808 (18) | 0.0578 (5) | |
| C2 | 0.50098 (15) | 0.3718 (3) | 0.64471 (19) | 0.0630 (6) | |
| H2 | 0.553081 | 0.358487 | 0.606754 | 0.076* | |
| C8 | 0.21562 (15) | 0.4992 (3) | 0.4654 (2) | 0.0704 (6) | |
| H8 | 0.260703 | 0.585579 | 0.448436 | 0.084* | |
| C7 | 0.23591 (14) | 0.3858 (3) | 0.56438 (18) | 0.0604 (5) | |
| C1 | 0.51178 (14) | 0.3912 (3) | 0.76354 (19) | 0.0565 (5) | |
| C12 | 0.16757 (15) | 0.2578 (3) | 0.5889 (2) | 0.0737 (6) | |
| H12 | 0.180146 | 0.181271 | 0.655006 | 0.088* | |
| C3 | 0.41015 (15) | 0.3728 (3) | 0.58389 (19) | 0.0642 (6) | |
| H3 | 0.401257 | 0.359785 | 0.503076 | 0.077* | |
| C11 | 0.08146 (16) | 0.2435 (3) | 0.5161 (2) | 0.0787 (7) | |
| H11 | 0.035605 | 0.158226 | 0.532243 | 0.094* | |
| C10 | 0.06496 (16) | 0.3583 (4) | 0.4192 (2) | 0.0709 (6) | |
| C9 | 0.13043 (16) | 0.4857 (3) | 0.3926 (2) | 0.0754 (6) | |
| H9 | 0.117272 | 0.561654 | 0.326283 | 0.090* | |
| H2B | 0.2854 (15) | 0.490 (3) | 0.8945 (17) | 0.106 (9)* | |
| H2A | 0.2140 (13) | 0.484 (4) | 0.780 (2) | 0.114 (10)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0876 (11) | 0.1025 (13) | 0.0647 (12) | 0.0106 (9) | −0.0035 (9) | −0.0071 (11) |
| N1 | 0.0702 (12) | 0.0638 (12) | 0.0744 (15) | 0.0050 (10) | 0.0068 (12) | −0.0060 (12) |
| O3 | 0.0786 (11) | 0.1494 (18) | 0.1005 (16) | −0.0237 (12) | 0.0033 (10) | −0.0310 (13) |
| O2 | 0.0715 (10) | 0.1330 (15) | 0.0988 (14) | −0.0058 (11) | 0.0202 (10) | −0.0176 (12) |
| N2 | 0.0719 (13) | 0.1179 (18) | 0.0591 (14) | −0.0019 (12) | 0.0092 (11) | −0.0076 (13) |
| N3 | 0.0814 (16) | 0.1079 (19) | 0.0760 (18) | 0.0001 (13) | 0.0000 (14) | −0.0284 (14) |
| O4 | 0.1077 (14) | 0.1460 (18) | 0.0828 (15) | 0.0023 (12) | −0.0234 (11) | 0.0033 (14) |
| C5 | 0.0680 (13) | 0.0557 (13) | 0.0501 (13) | −0.0050 (10) | 0.0135 (11) | −0.0001 (10) |
| C6 | 0.0709 (13) | 0.0582 (13) | 0.0481 (12) | −0.0029 (10) | 0.0045 (11) | 0.0007 (10) |
| C4 | 0.0708 (13) | 0.0515 (12) | 0.0512 (13) | −0.0050 (10) | 0.0102 (11) | 0.0016 (11) |
| C2 | 0.0746 (14) | 0.0591 (14) | 0.0573 (15) | −0.0005 (11) | 0.0171 (12) | −0.0026 (11) |
| C8 | 0.0774 (14) | 0.0767 (15) | 0.0542 (14) | −0.0036 (12) | 0.0025 (12) | 0.0066 (12) |
| C7 | 0.0715 (13) | 0.0594 (13) | 0.0490 (13) | −0.0049 (11) | 0.0063 (11) | −0.0043 (11) |
| C1 | 0.0645 (12) | 0.0457 (12) | 0.0580 (15) | 0.0001 (10) | 0.0063 (11) | −0.0007 (11) |
| C12 | 0.0873 (15) | 0.0735 (15) | 0.0578 (15) | −0.0149 (13) | 0.0047 (13) | 0.0043 (13) |
| C3 | 0.0862 (15) | 0.0601 (13) | 0.0468 (13) | −0.0011 (11) | 0.0123 (12) | 0.0004 (11) |
| C11 | 0.0838 (16) | 0.0794 (16) | 0.0702 (18) | −0.0193 (13) | 0.0045 (13) | −0.0057 (15) |
| C10 | 0.0702 (14) | 0.0817 (17) | 0.0562 (16) | 0.0010 (12) | −0.0033 (12) | −0.0183 (13) |
| C9 | 0.0838 (15) | 0.0808 (17) | 0.0577 (15) | 0.0032 (13) | 0.0001 (13) | 0.0054 (13) |
Geometric parameters (Å, º)
| O1—N1 | 1.228 (2) | C2—C1 | 1.369 (3) |
| N1—O2 | 1.214 (2) | C2—C3 | 1.372 (3) |
| N1—C1 | 1.466 (3) | C2—H2 | 0.9300 |
| O3—N3 | 1.209 (2) | C8—C9 | 1.367 (3) |
| N2—C5 | 1.375 (3) | C8—C7 | 1.389 (3) |
| N2—H2B | 0.922 (17) | C8—H8 | 0.9300 |
| N2—H2A | 0.894 (17) | C7—C12 | 1.392 (3) |
| N3—O4 | 1.224 (3) | C12—C11 | 1.378 (3) |
| N3—C10 | 1.477 (3) | C12—H12 | 0.9300 |
| C5—C6 | 1.390 (3) | C3—H3 | 0.9300 |
| C5—C4 | 1.408 (3) | C11—C10 | 1.373 (3) |
| C6—C1 | 1.370 (3) | C11—H11 | 0.9300 |
| C6—H6 | 0.9300 | C10—C9 | 1.368 (3) |
| C4—C3 | 1.389 (3) | C9—H9 | 0.9300 |
| C4—C7 | 1.482 (3) | ||
| O2—N1—O1 | 123.1 (2) | C7—C8—H8 | 119.5 |
| O2—N1—C1 | 118.3 (2) | C8—C7—C12 | 118.8 (2) |
| O1—N1—C1 | 118.63 (19) | C8—C7—C4 | 120.41 (19) |
| C5—N2—H2B | 115.9 (14) | C12—C7—C4 | 120.6 (2) |
| C5—N2—H2A | 119.6 (17) | C2—C1—C6 | 122.8 (2) |
| H2B—N2—H2A | 116 (2) | C2—C1—N1 | 119.0 (2) |
| O3—N3—O4 | 123.1 (2) | C6—C1—N1 | 118.2 (2) |
| O3—N3—C10 | 118.6 (3) | C11—C12—C7 | 120.5 (2) |
| O4—N3—C10 | 118.3 (3) | C11—C12—H12 | 119.7 |
| N2—C5—C6 | 119.4 (2) | C7—C12—H12 | 119.7 |
| N2—C5—C4 | 121.8 (2) | C2—C3—C4 | 122.7 (2) |
| C6—C5—C4 | 118.80 (19) | C2—C3—H3 | 118.6 |
| C1—C6—C5 | 119.9 (2) | C4—C3—H3 | 118.6 |
| C1—C6—H6 | 120.1 | C10—C11—C12 | 118.4 (2) |
| C5—C6—H6 | 120.1 | C10—C11—H11 | 120.8 |
| C3—C4—C5 | 118.49 (19) | C12—C11—H11 | 120.8 |
| C3—C4—C7 | 118.3 (2) | C9—C10—C11 | 122.6 (2) |
| C5—C4—C7 | 123.25 (18) | C9—C10—N3 | 119.0 (3) |
| C1—C2—C3 | 117.28 (19) | C11—C10—N3 | 118.4 (2) |
| C1—C2—H2 | 121.4 | C8—C9—C10 | 118.6 (2) |
| C3—C2—H2 | 121.4 | C8—C9—H9 | 120.7 |
| C9—C8—C7 | 121.0 (2) | C10—C9—H9 | 120.7 |
| C9—C8—H8 | 119.5 | ||
| N2—C5—C6—C1 | −179.0 (2) | O2—N1—C1—C6 | −170.96 (19) |
| C4—C5—C6—C1 | 1.3 (3) | O1—N1—C1—C6 | 9.7 (3) |
| N2—C5—C4—C3 | 177.7 (2) | C8—C7—C12—C11 | 0.2 (3) |
| C6—C5—C4—C3 | −2.6 (3) | C4—C7—C12—C11 | −175.9 (2) |
| N2—C5—C4—C7 | −2.1 (3) | C1—C2—C3—C4 | 0.0 (3) |
| C6—C5—C4—C7 | 177.60 (19) | C5—C4—C3—C2 | 2.0 (3) |
| C9—C8—C7—C12 | −0.3 (3) | C7—C4—C3—C2 | −178.2 (2) |
| C9—C8—C7—C4 | 175.8 (2) | C7—C12—C11—C10 | 0.0 (3) |
| C3—C4—C7—C8 | −50.6 (3) | C12—C11—C10—C9 | −0.1 (3) |
| C5—C4—C7—C8 | 129.1 (2) | C12—C11—C10—N3 | −179.4 (2) |
| C3—C4—C7—C12 | 125.4 (2) | O3—N3—C10—C9 | −175.5 (2) |
| C5—C4—C7—C12 | −54.9 (3) | O4—N3—C10—C9 | 3.9 (3) |
| C3—C2—C1—C6 | −1.4 (3) | O3—N3—C10—C11 | 3.8 (3) |
| C3—C2—C1—N1 | −179.90 (18) | O4—N3—C10—C11 | −176.8 (2) |
| C5—C6—C1—C2 | 0.7 (3) | C7—C8—C9—C10 | 0.2 (3) |
| C5—C6—C1—N1 | 179.24 (19) | C11—C10—C9—C8 | 0.0 (3) |
| O2—N1—C1—C2 | 7.6 (3) | N3—C10—C9—C8 | 179.3 (2) |
| O1—N1—C1—C2 | −171.76 (19) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H2B···O1i | 0.92 (2) | 2.36 (2) | 3.229 (3) | 157 (2) |
| N2—H2A···O4ii | 0.89 (2) | 2.50 (2) | 3.345 (3) | 157 (2) |
| C6—H6···O1i | 0.93 | 2.54 | 3.308 (3) | 140 |
| C9—H9···O3iii | 0.93 | 2.57 | 3.496 (3) | 174 |
Symmetry codes: (i) −x+1, −y+1, −z+2; (ii) −x, −y+1, −z+1; (iii) −x, y+1/2, −z+1/2.
References
- Bastiansen, O. & Samdal, S. (1985). J. Mol. Struct. 128, 115–125.
- Bastiansen, O. & Traetteberg, M. (1962). Tetrahedron, 17, 147–154.
- Boonstra, E. G. (1963). Acta Cryst. 16, 816–823.
- Brock, C. P. (1980). Acta Cryst. B36, 968–971.
- Brock, C. P. & Minton, P. (1989). J. Am. Chem. Soc. 111, 4586–4593.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Lehane, R. L., Golen, J. A., Rheingold, A. L. & Manke, D. R. (2014). Acta Cryst. E70, o305. [DOI] [PMC free article] [PubMed]
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Ol’khovik, V. K., Pap, A. A., Vasilevskii, V. A., Galinovskii, N. A. & Tereshko, S. N. (2008). Russ. J. Org. Chem. 44, 1172–1179.
- Sekine, A., Ohashi, Y., Yoshimura, K., Yagi, M. & Higuchi, J. (1994). Acta Cryst. C50, 1101–1104. [DOI] [PubMed]
- Shaikh, N. S., Parkin, S., Luthe, G. & Lehmler, H.-J. (2008). Chemosphere, 70, 1694–1698. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Stoe & Cie (2002). X-AREA and X-RED32. Stoe & Cie, Darmstadt, Germany.
- Sutherland, H. H. & Ali-Adib, Z. (1986). Acta Cryst. C42, 432–433.
- Trotter, J. (1961). Acta Cryst. 14, 1135–1140.
- Umeda, Y., Aiso, S., Yamazaki, K., Ohnishi, M., Arito, H., Nagano, K., Yamamoto, S. & Matsushima, T. (2005). J. Vet. Med. Sci. 67, 417–424. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, Global. DOI: 10.1107/S205698901700408X/su5355sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698901700408X/su5355Isup2.hkl
Supporting information file. DOI: 10.1107/S205698901700408X/su5355Isup3.cml
CCDC reference: 1537734
Additional supporting information: crystallographic information; 3D view; checkCIF report






