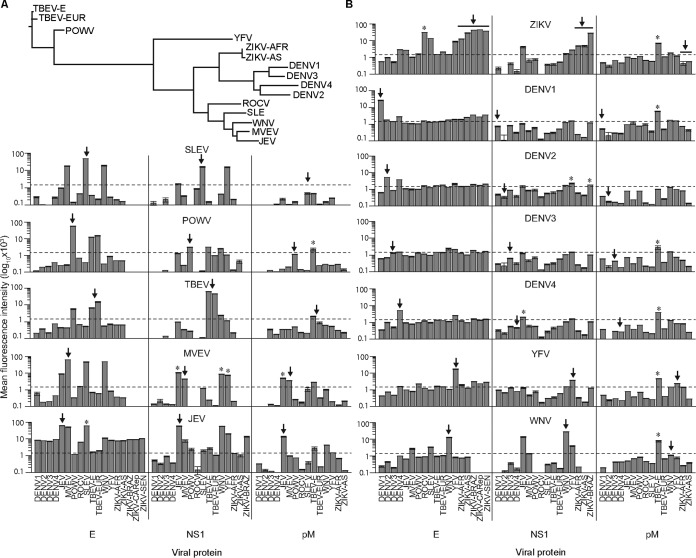
FIG 1.
Phylogenetic relationships and recognition of microarrayed antigens by virus-specific antibody standards. (A) The phylogenies of flaviviruses examined in this study were inferred from an alignment of amino acid sequences from envelope (E) proteins. (B) Microarrays of E, nonstructural protein 1 (NS1), and premembrane (pM) proteins probed with mouse polyclonal antibodies generated against each virus shown (centered labels above each row of bar graphs). Antibody binding data are shown as log10-transformed mean fluorescence intensities (±standard errors of the means [SEM] [error bars]), and the arrows indicate the virus-specific antigens. Heterologous antigens that exhibit increased recognition compared to the virus-specific antigen are labeled with an asterisk (P < 0.05, one-way ANOVA with Tukey's range test). Virus abbreviations: YFV, yellow fever virus; SLEV, St. Louis encephalitis virus; DENV, dengue virus; DENV1, dengue virus serotype 1; POWV, Powassan virus; TBEV-E, tick-borne encephalitis virus, Eastern strain; TBEV-EUR, tick-borne encephalitis virus, European strain; MVEV, Murray Valley encephalitis virus; WNV, West Nile virus; ZIKV, Zika virus; ZIKV-AFR, ZIKV from Africa; ZIKV-AS, ZIKV from Asia; JEV, Japanese encephalitis virus; ROCV, Rocio virus.