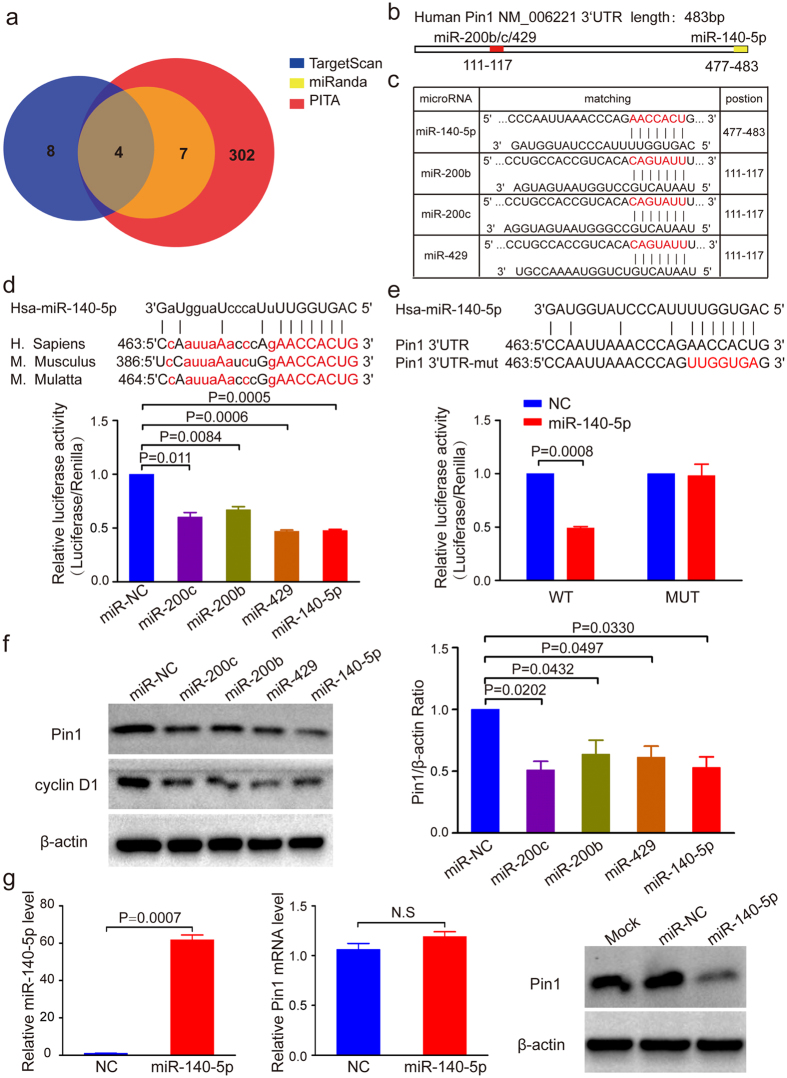
Figure 1. Pin1 is a direct target of miR-140-5p in HCC.
(a) Bioinformatic prediction of candidate miRNAs targeting the Pin1 3′UTR. (b) Schematic diagram showing the predicted binding region of miRNAs in Pin1 3′UTR. (c) The predicted binding sites for miR-140-5p in Pin1 3′UTR. The red nucleotides are the seed-pairing target sites of miRNAs. (d) Bioinformatic analyses show Pin1 as a promising target of miR-140-5p (top panel) and miR-140-5p reduces Pin1 expression (bottom panel), as assayed by a luciferase reporter. The seed sequences of miR-140-5p within the Pin1 3′UTR are evolutionarily highly conserved across mammals as marked red. Capitalized letters are the conserved binding sites that directly interact with miR-140-5p. Dual-luciferase assay showed that miR-140-5p and miR-200s reduce luciferase activity by about 50%. (e) MiR-140-5p targets wild-type Pin1 3′UTR, but not its mutant. Luciferase constructs bearing a Pin1 3′UTR (WT) or Pin1 3′UTR containing mutated binding sequences of miR-140-5p (Mut) were cotransfected with miR-140-5p. Results showed that miR-140-5p reduces luciferase activity by 50%, but that was abolished when miR-140-5p binding sequences on Pin1 3′UTR was mutated. (f) MiR-140-5p downregulates Pin1 and cyclin D1 expression, as detected by Western blot analysis. β-actin served as loading control. (g) MiR-140-5p downregulates Pin1 at the translational level. Huh7 cells were infected with lentiviruses expressing miR-140-5p followed by selection with indicated concentration of puromycin. The expression of miR-140-5p and Pin1 protein were significantly increased and decreased, respectively, while Pin1 mRNA level had no significant change. In all panels, bar graphs represent mean ± SEM of three independent experiments. The statistical significance of all tests was accepted for P < 0.05.