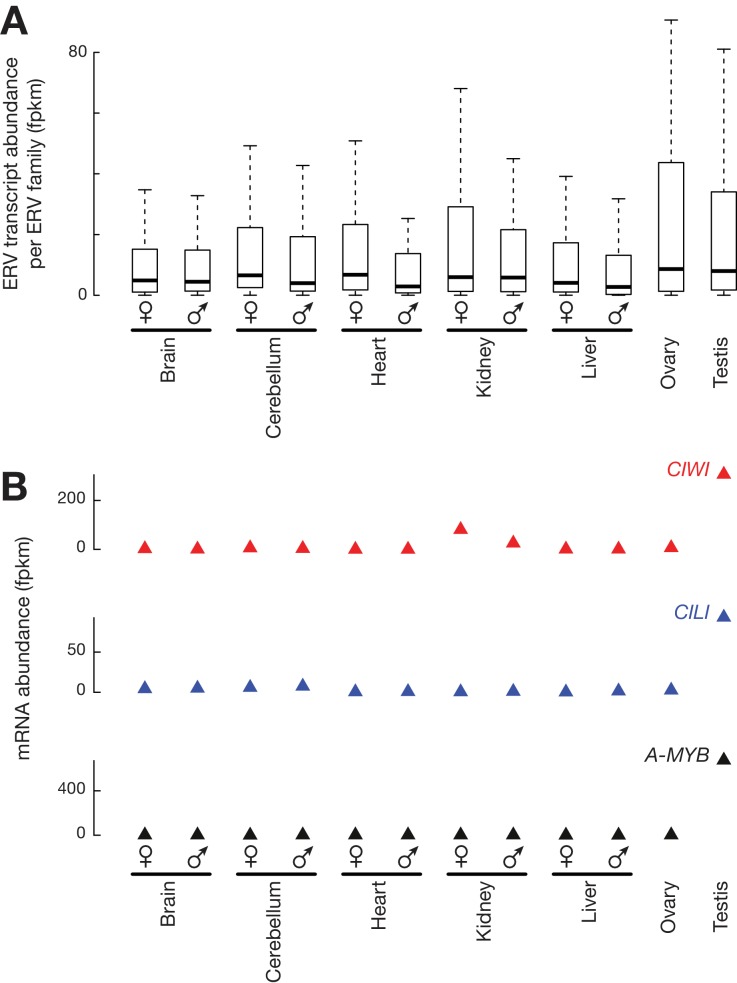
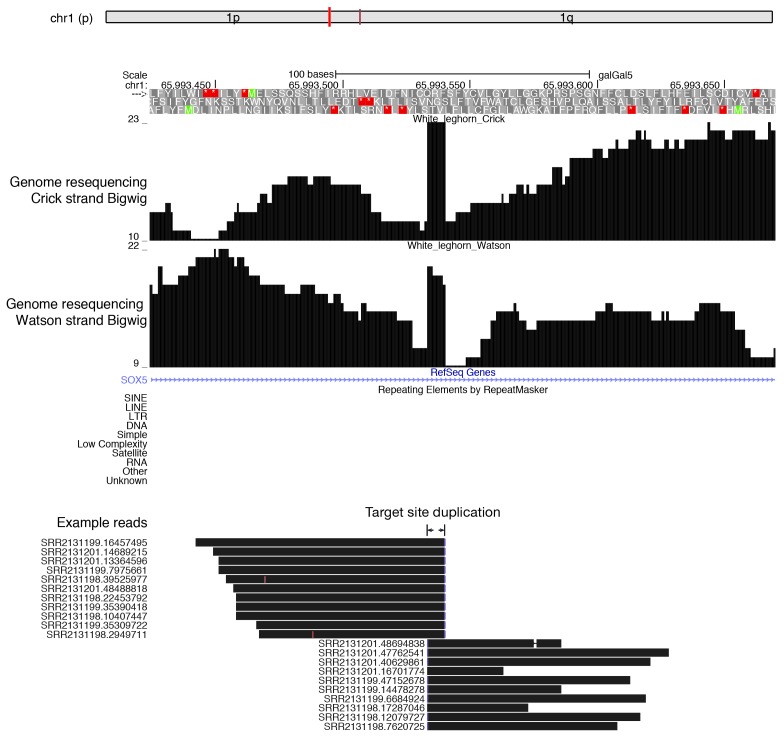
Figure 1. Active ERVs in White Leghorn testis.
(A) A254 absorbance profile of 10% to 50% sucrose density gradients of testis lysates from adult rooster. From top to bottom, plots show the relative abundance of 18S rRNA, β-ACTIN mRNA, CILI mRNA, CIWI mRNA, chicken Malat1 lncRNA, CR1B, CR1F, EAV-HP, and ALVE quantified by RT-qPCR. Data were normalized to a spike-in control RNA. (B) Scatter plots of transcript abundance versus ribosome density. Each black dot represents an mRNA expressed in testis. Each filled red circle represents an ERV family, and each open red circle represents any other TE family, including DNA transposons and CR1 superfamily; rpkm, reads per kilobase of transcript per million mapped reads; fpkm, fragments per kilobase of transcript per million mapped reads. (C) Normalized reads of White Leghorn RPFs (Top), White Leghorn piRNAs (Middle), and Red Jungle Fowl small RNA reads (>23 nt) (Bottom). Blue represents sense mapping reads; Red represents anti-sense mapping reads. The gene organization of ALVE is also shown. Gag, group-specific antigen; Pol, polymerase; Env, envelope protein; ppm, parts per million. (D) Circos plot representing the locations, from periphery to center, of cytological position (black lines represent centromeres), piRNA clusters in White Leghorn (Black lines represent conserved piRNA clusters; White lines represent divergent piRNA clusters), putative new insertions discovered by TEMP (tiles) using genomic resequencing of White Leghorn, and 2 × 2 contingency table for Fisher’s exact test to assess the significance of the coincidence of transcription and translation of each TE family. The table data correspond to the number of TE families in each category and, in parentheses, the number of TE families in each category with recent transpositions.