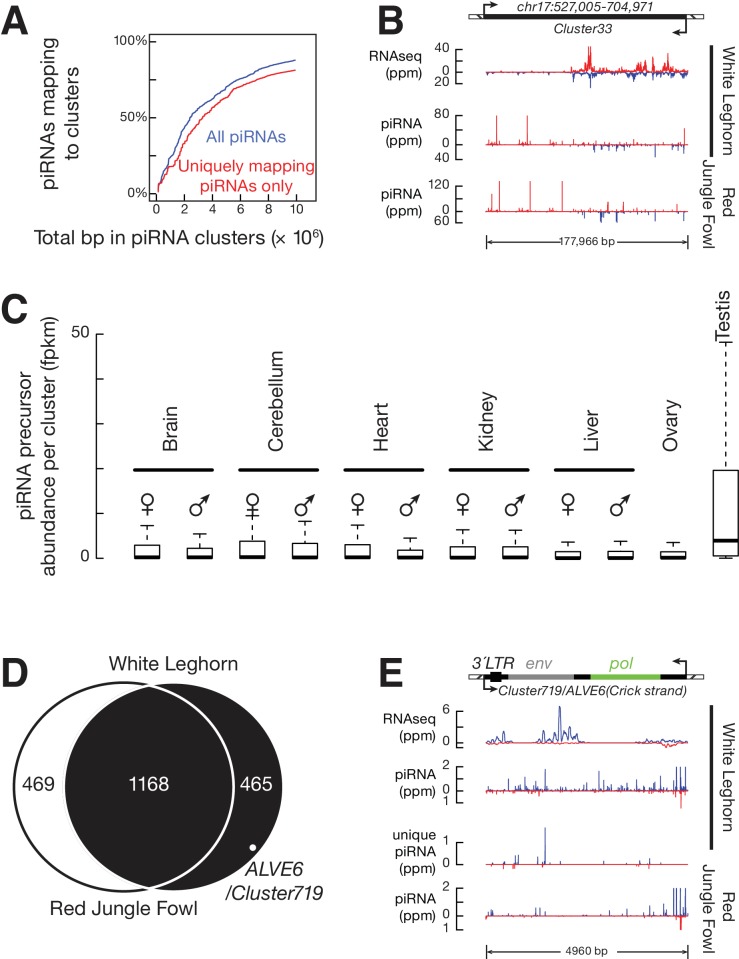
Figure 4. ALVE6 is the primary piRNA-producing locus for viral piRNAs.
(A) Cumulative distributions for all piRNAs (Blue) and for uniquely mapping piRNAs (Red) on the 1633 piRNA loci in White Leghorn. (B) An example of conserved piRNA-producing loci, Cluster33, in chicken. Normalized RNA-seq reads of piRNA precursors, piRNAs in White Leghorn, and piRNAs in Red Jungle Fowl. Blue represents Watson-strand piRNAs; Red represents Crick-strand piRNAs. (C) Box plots showing abundance of piRNA precursors in 12 chicken tissues. (D) Venn diagram showing piRNA clusters defined in Red Jungle Fowl (White) and White Leghorn (Black). (E) Normalized RNA-seq reads of piRNA precursors in White Leghorn, White Leghorn piRNAs, unique mapping piRNAs, and Red Jungle Fowl small RNA reads (>23 nt).