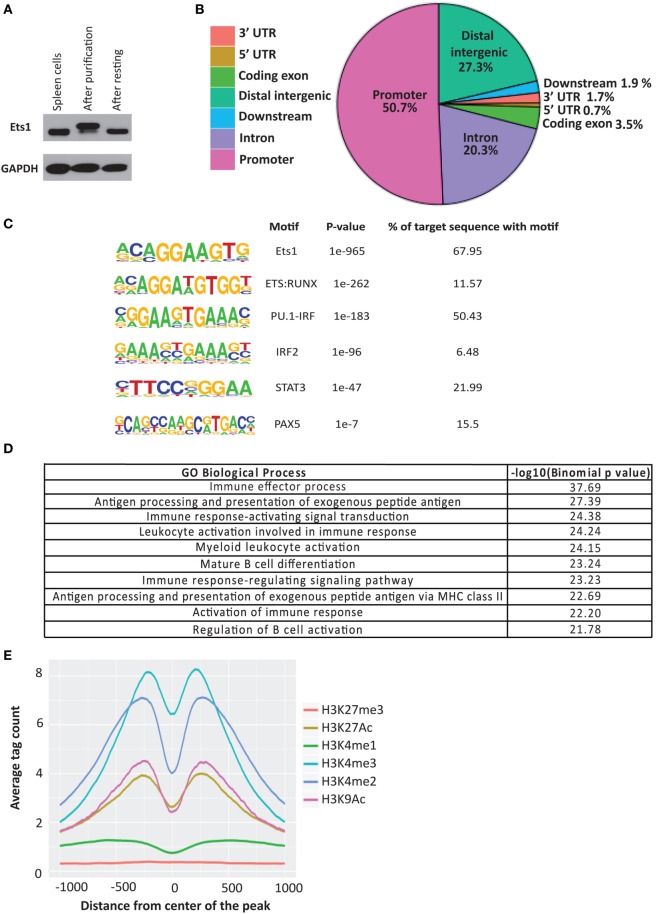
Figure 1.
Identification of Ets1-binding sites in mouse B cells. (A) Western blot to show phosphorylation of Ets1 in freshly isolated B cells versus rested B cells. GAPDH serves as loading control. (B) Pie chart of location of Ets1 sites in the genome. (C) Motifs enriched in Ets1-bound regions. Shown are overrepresented transcription factor-binding motifs localized in the Ets1 peaks and the percent of sites with that motif. (D) Gene ontology biological terms associated with Ets1-binding peaks in B cells. (E) Analysis of epigenetic features surrounding Ets1-bound regions by mapping adjacent histone modifications. Data come from the ENCODE Consortium or from the studies described in Ref. (50, 51).