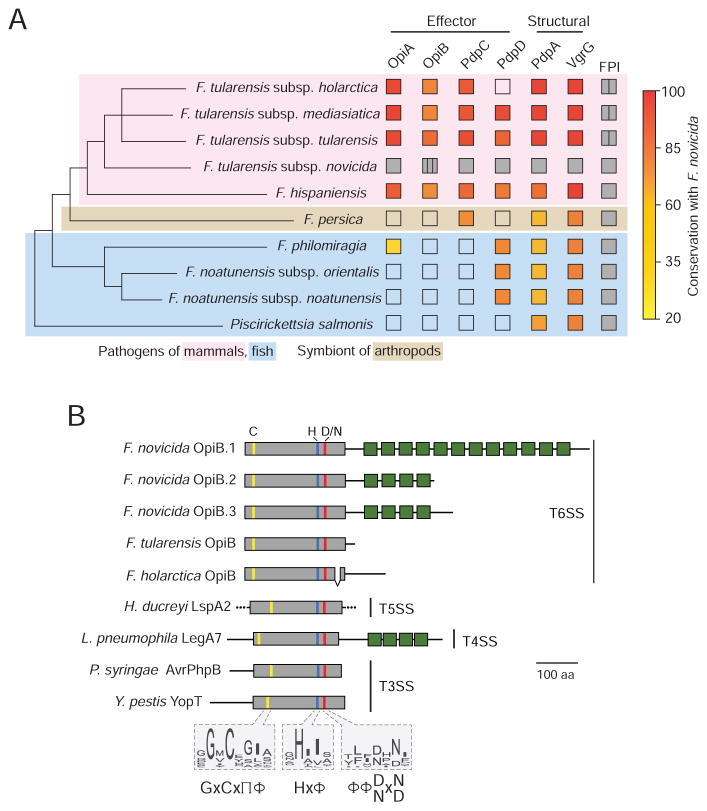
Figure 6. OpiA and OpiB are Virulence-associated Proteins.
(A) Phylogenetic tree of Francisella and closely related organisms that possess the FPI (see methods) indicating the presence (filled) or absence (open) of secreted F. novicida T6SSii effectors and structural proteins identified in this study. If present, the degree of conservation with the F. novicida ortholog is indicated (fill color). The tree is adapted from Sjodin et al.; branch lengths do not represent evolutionary distance (Sjodin et al., 2012). Vertical lines within boxes indicate multiple copies of a given element and background colors denote host-specificities as indicated.
(B) Overview of OpiB domain organization and conservation of its predicted N-terminal protease domain among diverse effector proteins. All proteins are shown to scale. The location of predicted or experimentally determined catalytic residues are indicated by colored lines, and box fill denotes protease domain (grey) and ankyrin motifs (green). Conserved catalytic motifs based on the alignment of a broader group of representative orthologs are shown as sequence logos. Amino acid composition at each position is shown according to (Aasland et al., 2002). The experimentally determined or predicted export pathway of each protein is provided at right. Proteins shown are: OpiB-1 A0Q6U1, OpiB-2 A0Q6U2, OpiB-3 A0Q6U3, OpiBFT Q5NH59, OpiBLVS Q2A3V2, LspA2 Q9ZHL3, LegA7 Q5ZYG9, AvrPhpB Q9F3T4, YopT O68703.