Abstract
In spite of the growing body of evidence on the biology of the Zebrafish embryo and stem cells, including the use of Stem Cell Differentiation Stage Factors (SCDSFs) taken from Zebrafish embryo to impact cancer cell dynamics, comparatively little is known about the possibility to use these factors to modulate the homeostasis of normal human stem cells or to modulate the behavior of cells involved in different pathological conditions. In the present review we recall in a synthetic way the most important researches about the use of SCDSFs in reprogramming cancer cells and in modulating the high speed of multiplication of keratinocytes which is characteristic of some pathological diseases like psoriasis. Moreover we add here the results about the capability of SCDSFs in modulating the homeostasis of human adipose-derived stem cells (hASCs) isolated from a fat tissue obtained with a novel-non enzymatic method and device. In addition we report the data not yet published about a first protein analysis of the SCDSFs and about their role in a pathological condition like neurodegeneration.
Keywords: Stem cell differentiation stage factors, cancer stem cells, human adipose-derived stem cells, cell reprogramming, cancer therapies, psoriasis, anti-aging treatments, neurodegeneration
Introduction
Current medical literature acknowledges that embryonic microenvironment is able to suppress tumor development during cell differentiating processes [1, 2]. Administration of carcinogenic substances during organogenesis leads in fact to embryonic malformations, but not to offspring tumor growth. However, administration of carcinogenic substances after complete organogenesis causes a rise in offspring tumor development [3-5]. These data indicate that cancer can be considered as a deviation in normal development that can be controlled by factors in embryonic microenvironment during the differentiating stages. Furthermore, it has been demonstrated that teratoma differentiates into normal tissues once implanted in the embryo [6].
Recently, it has been shown that implantation of melanoma cells into Zebrafish embryos does not result in tumor development, while in the adult animal, a tumor is formed [7]. Moreover, injection of melanoma cells in Zebrafish extra-embryonic membranes originated Zebrafish neuronal cells. This demonstrates that cancer cells can differentiate in normal tissues when implanted in embryos [8]. In addition, it was demonstrated that other tumors, including leukemia, liver and breast tumor cells, can differentiate into normal tissue when implanted in the embryo [9-12].
The term “reprogramming” was initially introduced to identify the transformation of a normal adult somatic cell into an embryonic-like stem cell, the induced pluripotent stem cells (iPS). The issue of cell reprogramming has now been extended to cancer (stem) cells to define any genetic or epigenetic intervention aimed at inducing differentiation of these cells into a normal phenotype and/or forcing them to become terminally differentiating cells. These interventions focus on the role of the embryonic microenvironment in tumor reprogramming. Intriguingly, it is now evident that the molecular mechanisms underlying normal stem cell differentiation and embryonic development do not stop after birth but are still in part operating and remodeled throughout the adult life to maintain the self-identity and the interplay between tissues and organs. To this end, it has been shown that the transcription factor GATA4 is a crucial regulator of both embryonic and postnatal heart development and morphogenic maintenance due to a fine tuning of its structural/regulatory domains [13]. Whereas the N-terminal domain of GATA4 is needed for promoting postnatal cardiomyocyte survival and for inducing cardiogenesis, other distinct residues and domains therein are crucial to mediate these effects [13]. A noteworthy example of morphogenetic flexibility is also provided by the existence of reverse pathways of transformation, from the postnatal stage back to an embryonic-like condition retaining the memory ability to re-differentiate backward to the same original phenotype. A vivid example of such flexibility is shown by the ability of post-natal cardiomyocytes to generate iPS cells with increased capacity toward cardiomyogenic re-differentiation [14]. Similarly, adult neurogenesis, a process of generating functional neurons from adult neural precursors, has been shown to occur throughout life in restricted brain regions in mammals, including the dentate gyrus of the hippocampus, the subventricular zone of the lateral ventricle, and the rostral migratory stream to the olfactory bulb [15]. This discovery is currently boosting emerging principles that have significant implications not only in stem cell biology, developmental neurobiology, and neural plasticity, but, remarkably, in disease mechanisms, including neurodegeneration.
Hence, a kind of memory/projection of the embryonic patterning may be conceived as a relevant background in tissue resident stem cells in the adulthood for the execution of self-healing and “learning” (acquirement of new knowledge) tasks. In this frame, degenerative diseases occurring in any organ (i.e. neurodegenerative diseases) may be viewed as a deviation from the normal potential of tissue resident stem cells to afford self-healing duties and the maintenance of tissue organ identity.
Akin to this perception, here we review several of our experimental findings over the past 20 years on the possibility to reprogram cancer cells in vitro as well as in vivo. In fact, we present results from controlled clinical studies on hepatocellular carcinoma at intermediate-advanced stage based on the treatment with Zebrafish Stem Cell Differentiation Stage Factors (SCDSFs) taken during precise stages of stem cell differentiating processes [16, 17]. We also report on our recent finding that the same SCDSFs obtained at early developmental stages acted as a major controller of stemness and senescence patterning in human adult adipose-derived stem cells [18]. Consistent with the concept of considering tissue degeneration a “flexible” deviation from a tissue identity program still entangled with embryogenetic memory, we show our recent findings on the ability of SCDSFs to prevent neurodegeneration in hippocampal cells of CA1 area in mice. Compounding the spectrum of exploitation of SCDSF potential for (stem) cell reprogramming, we recently succeeded in using Zebrafish embryo differentiation factors to reduce keratosis and ameliorate symptoms in patients affected by psoriasis [19-21], a T cell-dependent immune-mediated disease of the skin and joints. Such result is also rewarding due to (i) the recent detection of functional circadian clocks in most, if not all, of skin cell types, (ii) the emergence of a close involvement of these circadian clocks in the control of UVB-induced DNA damage and skin cancers, and (iii) the implication for the targeted modulation of stem cell-mediated immunomodulatory action and control of aging processes [22, 23].
Role of SCDSFS in cancer cell lines and in mice carcinoma cells
In vitro effects of SCDSFs on different human tumor cell lines have been investigated in a number of studies [24-28]. Seven different human tumor cell lines were treated with factors taken from Zebrafish embryos at different developmental phases, specific of the beginning, intermediate and final embryonic differentiation stages. In general, a reduced growth rate was seen when tumor cells lines were treated with factors drawn during the different developmental stages, ranging from 73% reduction for the glioblastoma cells to 26% for the melanoma cells. No proliferative effects have been reported, except from a weak tumoral growth with factors extracted at a very early stage of embryonic development in which the differentiation processes did not begin, like morula stage. These data confirm the intuition that in the embryo, during the differentiating stages, there are networks of factors able to readdress tumoral cells towards a normal path. Those networks appear in the very first phases of the gastrulation, while they are absent in merely multiplicative stages [24].
Several studies were carried out in order to unravel the molecular mechanisms involved in tumor growth inhibition mediated by Zebrafish embryonic extracts, showing that molecules that have a fundamental role in regulation of the cell cycle, such as p53 and retinoblastoma protein (pRb) were affected. More precisely, a p53 transcriptional regulation took place, highlighted by a considerable increase of the p53 protein expression in some of the tumor cell lines, such as the glioblastoma multiforme and the melanoma [25]. In other tumor cell lines, such as kidney adenocarcinoma, the growth reduction was due to changes in phosphorylation of pRb [26], which is known to regulate transcription of E2F-1 and thereby controlling the cell cycle.
Moreover, apoptotic events as well as cell differentiation events were studied, in order to understand the consequences of cell cycle regulation in tumor cells induced by differentiation factors. The analysis was carried out on colon adenocarcinoma cells, showing activation of an apoptotic pathway dependent on p73, as well as an increase in the cell differentiation marker e-cadherin [27].
Finally, in order to ascertain if SCDSFs could synergistically/additively interact with 5-Fluorouracil (5-Fu), whole cell-count, flow-cytometry analysis and apoptotic parameters were recorded in human colon cancer cells (Caco-2) treated with SCDSFs 3 μg/ml in association or not with 5-Fu in the sub-pharmacological therapeutic range (0.01 mg/ml). Cell proliferation was significantly reduced by SCDSFs, meanwhile SCDSF+5-Fu leads to an almost complete growth-inhibition. SCDSFs produce a significant apoptotic effect, and the association with 5-Fu leads to an enhanced additive apoptotic rate at both 24 and 72 hours. SCDSFs alone and in association with 5-Fu trigger both the extrinsic and the intrinsic apoptotic pathways, activating caspase-8, -3 and -7. These data suggest that Zebrafish embryonic factors could improve chemotherapy efficacy by reducing anti-apoptotic proteins involved in drug-resistance processes [28]. Therefore, the molecular mechanisms underlying the tumor growth reduction seen after treatment with SCDSFs can be summarized as follows: the cell cycle stops in G1-S or G2-M phase, according to the tumor type, genetic damage repair and cell re-differentiation, or tumor cells apoptosis if reparation is not possible because of mutation gravity.
The effects of SCDSFs on tumor growth were also observed in vivo after subcutaneous injection of primary Lewis Lung Carcinoma cells into C57BL/6 female syngenic mice weighing 18-20 gr. A single cell suspension of tumor cells was prepared by mechanical dissociation of tumor mass: 50 μL of Dulbecco phosphate buffered saline (DPBS) containing 106 viable tumor cells were mixed with SCDSFs and used in the treated animals, while the control group received 50 μL of DPBS. A highly significant difference was noted (p<0.001) between treated and control mice both in terms of primary tumor development and of the survival rate in favor of the treated mice [29].
SCDSFS in clinical trials on intermediate-advanced hepatocellular carcinoma (HCC)
From January the 1st 2001 to April the 31st 2004 a randomized controlled clinical trial was conducted on 179 patients affected by hcc in an intermediate-advanced stage. Since no further treatments were possible, a product fine tuned on the basis of above mentioned studies was administrated. The posology was 30 sublingual drops of the Zebrafish embryo differentiation factors three times a day. The sublingual solution was chosen because the composition of the active fraction is composed of low molecular weight proteins (see the data about the protein analysis of SCDSFs).
Objective tumor response, overall survival and performance status have been evaluated. Results showed that 19.8% of the patients experienced a regression and 16% experienced a stabilization with an overall survival of more than 60% of the responsive patients after 40 months, compared to 10% of the non responsive patients.
A wide improvement of performance status has been registered in a great majority of patients (82.6%), also in those who experienced a progression of the disease [16]. A more recent study confirms the role of SCDSFs in determining complete response in primitive intermediate advanced liver cancer in 13.1% patients [17].
SCDSFS in human adipose-derived stem cells (HASCS)
The possibility to address the fate of hASCs, isolated from a fat tissue obtained with a novel non-enzymatic method and device (Lipogems) [30], was explored by exposing them to SCDSFs [18].
SCDSFs taken during the late developmental stages (20 somites and pharyngula stages) reduced cell viability and elicited caspase-3 mediated apoptosis. This effect did not include Bax or Bcl-2 transcription. This circumstance has long been observed, as shown in the case of Bax-independent, caspase-3-related apoptosis induced by hepatocyte growth factor (HGF) in rat liver epithelial cells and recently confirmed in both normal and cancer cells [31].
Unlike SCDSFs taken during the late developmental stages, SCDSFs taken during the early developmental stage (50% epiboly stage) did not induce hASC apoptosis, nor did it decrease cell viability. Indeed, SCDSFs of the early developmental stage were able to regulate the stem cell expression of multipotency, enhancing the stemness genes Oct-4, Sox-2 and c-Myc. In addition to affecting stemness genes which maintain stem cell identity [32], SCDSFs also elicited transcriptional activation of two major mechanisms capable of opposing stem cell senescence, including the gene expression of TERT, the catalytic subunit of telomerase, and the transcription of Bmi-1. This is a member of the Polycomb and Trithorax families of repressors which acts as essential factors for self-renewal of adult stem cells, and as a key telomerase independent repressor of cell aging [33].
Thus, this study showed that human stem cell exposure to SCDSFs taken during the early developmental stage of Zebrafish embryo may represent a very effective tool to increase stem cell expression of multipotency and promote both telomerase-dependent and -independent antagonists of cell senescence. On the contrary SCDSFs taken during the late developmental stages decrease cell viability and address cells toward senescence. This strategy did not require any gene manipulation through viral vector mediated gene transfer, or expensive synthetic chemistry. These results show for the first time that it is possible to address human mesenchymal stem cells towards different and opposite directions, tuning in specific, physiological way the regulation of different genes.
Neuroprotective role of SDCSFS
We present here, for the first time, some recent findings on the ability of SCDSFs to prevent neurodegeneration in hippocampal cells of CA1 area in mice.
In order to evaluate the neuroprotective effect of SCDSFs, murine hippocampal slices of the CA1 area were prepared and cultured as described by Gardoni et al. [34] and four Zebrafish embryo solutions were prepared as follows: A (50% epiboly plus tail bud stage extracts), B (5 somites stage), C (20 somites plus pharingula stage) and Mix ABC (a mixture of the three solutions A, B and C) [24].
Organotypic hippocampal slices were treated with N-Methyl-D-Aspartate (NMDA) 50 μM and 300 μM for 1 hour to induce mortality and a propidium iodide (PI) coloration was performed after 24 hours [35]. After fixing, the CA1 area was acquired and mortality was analyzed considering the average PI-fluorescence intensity using as a term of comparison the maximum cell damage obtained by exposing the organotypic slices to NMDA.
We first observed that treatment with NMDA 50 μM and 300 μM induced an increase of mortality in both kinds of the treatments compared with the controls (p=0.002 and p=0.0002 respectively).
Then we evaluated the neuroprotective effect of SCDSFs after treatment with three toxic stimuli administered for 1 hour at the 14th day of culture: they were serum deprivation, NMDA 50 μM and NMDA 300 μM. Analyses were performed after 24 hours from treatments.
We noticed that treatment with the Mix ABC (dilution 1:100) administered together with each of the three toxic stimuli reduced in a significant manner the neuronal mortality caused by both serum deprivation and NMDA treatments.
In fact SCDSFs significantly reduce the neuronal mortality caused by serum deprivation (-31,6 ± 6,2%, p=0.005) as shown in (Fig. 1).
Fig. (1).
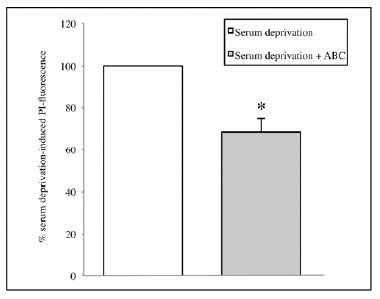
The effect of the mix ABC on CA1 area cell mortality after 1 hour of serum deprivation (*p=0.005).
Moreover, treatment with NMDA 50 μM significantly increases cell mortality compared with the controls (p=0.002) and SCDSFs significantly reduce the neuronal mortality caused by NMDA 50 μM treatment (p=0.01, Fig. 2).
Fig. (2).
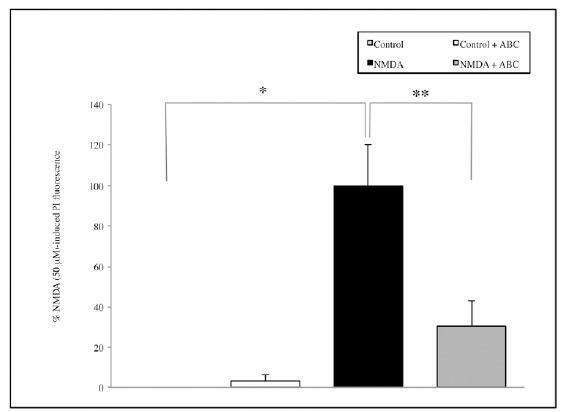
The effect of the mix ABC on CA1 area cell mortality after 1 hour NMDA 50 μM treatment (*p=0.002; **p=0.01).
Similarly, treatment with NMDA 300 μM significantly increases cell mortality compared with the controls (p=0.0002) and SCDSFs significantly reduce the neuronal mortality caused by NMDA 300 μM treatment (p=0.009, Fig. 3).
Fig. (3).
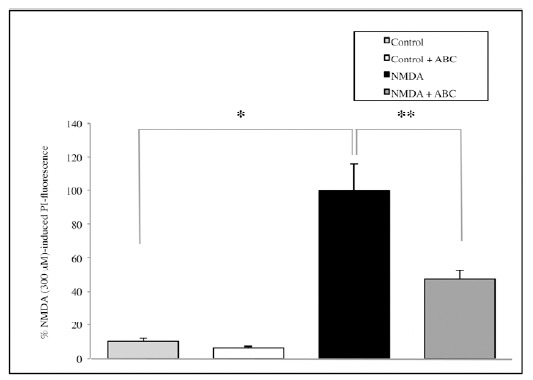
The effect of the mix ABC on CA1 area cell mortality after 1 hour NMDA 300 μM treatment (*p=0.0002; **p=0.009).
Subsequently, the potential neuroprotective activities of A or B or C were investigated. Also in this case, the experiments showed a reduction in mortality, overall for A extract but results are not enough significant, neither in the serum deprivation group (Fig. 4) nor in the NMDA group (Fig. 5). Thus, the whole informational set with a redundance of differentiation stage factors is needed to produce an effective result.
Fig. (4).
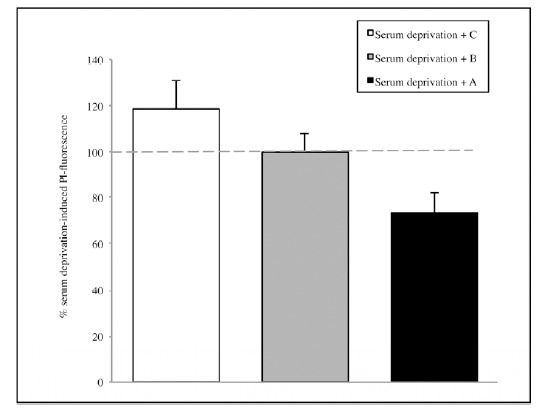
The effect of the single solutions A, B and C on CA1 area of hippocamp after 1 hour of serum deprivation. Values are expressed as percentage of samples treated with serum deprivation without SCDSFs.
Fig. (5).
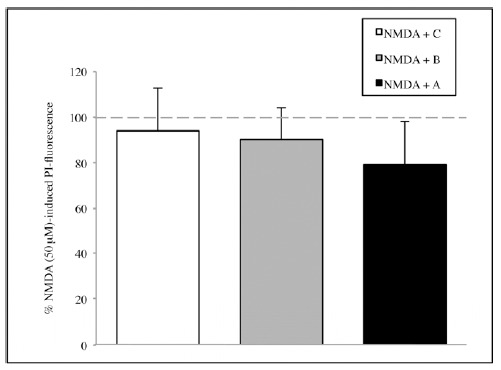
The effect of the single solutions A, B and C on CA1 area of hippocamp after 1 hour of NMDA 50 μM treatment. Values are expressed as percentage of samples treated with NMDA 50 μM without SCDSFs.
Experimental research and clinical studies on psoriasis
It was also investigated the anti-proliferative effects of SCDSFs by addressing the mitochondrial function (MTT assay) and cell nuclei distribution (Hoechst staining) in epidermal cell cultures stimulated with fetal calf serum (FCS) or epidermal growth factor (EGF). SCDSFs significantly inhibited cell proliferation induced by either approach, although the effect was stronger in cells stimulated with FCS [36]. Three clinical trials were conducted to evaluate the efficacy in cases of psoriasis following the administration of a mix of all 5 Zebrafish embryo developmental stage extracts added with Boswelia serrata, 18-beta glicirretic acid, Zanthoxylum alatum, 7-deidro-cholesterol and vitamin E. Results show 80% clinical objective improvements, with a reduction of keratosis and itch after 20-30 days from the beginning of the treatment [19-21].
Protein analysis of SCDSFS
To better know the content of the SCDSFs that we employed for our researches, we begun to perform protein analysis of the extracts, and here we present our first results.
Firstly, protein content of the five Zebrafish embryo extracts resuspended in a glycero-alcoholic solution [18, 24] was analyzed on an one-dimensional Sodium Dodecyl Sulphate - PolyAcrylamide Gel Electrophoresis (SDS-PAGE) [37]. After Coomassie staining [38], the protein amount was evaluated as pixel intensity and relative abundances were expressed as percentage of the total intensity. As shown in Fig. 6, in all five extracts, three main protein clusters are distinguishable according to their molecular weight, i.e. over 45 kDa, around 25-35 kDa and less than 20 kDa. Anyway, the relative protein abundance is different among the five samples.
Fig. (6).
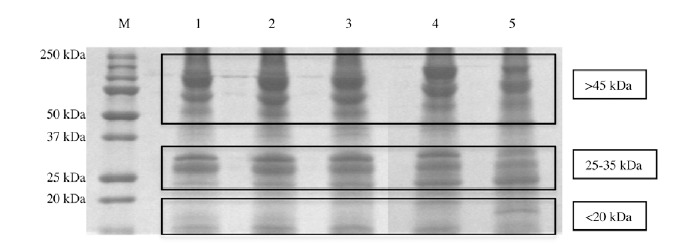
Representative 12% SDS-PAGE gel of Zebrafish embryo extracts resuspended in a glycero-alcoholic solution. Lanes: M) Broad-range protein molecular weight markers (in kDa); 1) 50% epiboly stage proteins; 2) tail bud stage proteins; 3) 5 somites stage proteins; 4) 20 somites stage proteins; 5) pharingula stage proteins.
At the beginning of the gastrula period (50% epiboly stage, Lane 1), the higher molecular weight cluster (> 45 kDa) represents the 45,8% of the bands intensity; this relative abundance is quite stable at the end of the gastrula period (tail bud stage, Lane 2) with a peack at the beginning of the segmentation, 46,1% (5 somites, Lane 3), while at the middle-late segmentation (20 somites, Lane 4 and pharyngula, Lane 5) this percentage composition decreases until the 43,9%.
The 25-35 kDa protein cluster abundance is quite stable in the gastrula period (Lanes 1 and 2), around 25.5%, while during the segmentation (Lanes 3, 4 and 5) it decreases until the 22.6%. At the beginning of the gastrula period (50% epiboly stage, Lane 1) the lowest molecular weight cluster (less then 20 kDa) represent the 28,5%; the cluster abundance is quite similar among the end gastrulation and early segmentation (Lanes 2 and 3) (29,4%) while at the end of the gastrulation stages (20 somites, Lane 4, and pharingula, Lane 5) this percentage composition increases until the 43,9%.
Then, all the proteins extracted from the earliest Zebrafish developmental investigated stage (50% epiboly) were identified by using a liquid chromatography–mass spectrometry (LC-MS/MS) analysis, after the in-gel digestion procedure as described by Della Corte and coll [39]. We listed in Table 1 the identified proteins with the correspondent NCBI accession number, the score, their isoelectric point (pI). Individual ions scores >36 indicate identity or extensive homology (p<0.05). Identified proteins include multiple form of yolk protein vitellogenin, heat shock protein (e.g. HSP8 and HSP70) and other proteins that have not been described before (indicated in Table 1 with an asterisk) [40, 41]. These proteins are implicated in many pathways as in signalling, cell cycle regulation, protein trafficking, chaperoning, protein synthesis and degradation.
Table 1.
List of proteins identified using the nano LC-ESI-Q-TOF with the specification of their NCBI accession number, name, score, molecular weight (MW) in Dalton (Da), isoelectric point (pI) and percentage sequence coverage. Proteins highlighted with asterisk (*) were not described before in Zebrafish embryo.
| Accession | Protein Name | Score | MW (Da) | pI | Coverage % |
|---|---|---|---|---|---|
| gi|166795887 | Vitellogenin 1 precursor | 1108 | 150308 | 8.68 | 19 |
| gi|94733730 | Vitellogenin 1 | 1039 | 149825 | 8.74 | 21 |
| gi|94733733 | Novel protein similar to vitellogenin 1 (vg1) | 913 | 149828 | 8.92 | 19 |
| gi|94733734 | Novel protein similar to vitellogenin 1 (vg1) | 835 | 150550 | 8.83 | 16 |
| gi|145337918 | Vtg1 protein | 780 | 116965 | 9.07 | 18 |
| gi|94733731 | Novel protein similar to vitellogenin 1 (vg1) | 762 | 149911 | 8.84 | 19 |
| gi|94732723 | Novel protein similar to vitellogenin 1 (vg1) | 745 | 147826 | 8.73 | 17 |
| gi|159155252* | Zgc:136383 protein | 720 | 124413 | 8.78 | 17 |
| gi|68448530 | Vitellogenin 5 | 559 | 149609 | 8.77 | 13 |
| gi|92097636 | Zgc:136383 | 402 | 28924 | 9.33 | 36 |
| gi|63100501 | Vtg1 protein | 345 | 36580 | 9.23 | 28 |
| gi|57864789 | Vitellogenin 7 | 341 | 24490 | 8.37 | 40 |
| gi|57864783 | Vitellogenin 4 | 334 | 31304 | 9.48 | 27 |
| gi|113678458 | Vitellogenin 2 isoform 1 precursor | 323 | 181208 | 8.70 | 11 |
| gi|125857991 | Zgc:136383 protein | 171 | 149328 | 8.93 | 9 |
| gi|15209312* | Procollagen type I alpha 2 chain | 169 | 147826 | 9.35 | 4 |
| gi|57864779 | Vitellogenin 2 | 122 | 69906 | 7.84 | 8 |
| gi|11118642 | Vitellogenin 3 precursor | 117 | 140477 | 6.92 | 2 |
| gi|303227889 | Vitellogenin 6 | 73 | 151677 | 8.84 | 4 |
| gi|13242157 * | Egg envelope protein ZP2 variant A | 71 | 48194 | 6.04 | 5 |
| gi|6644111 * | Nucleoside diphosphate kinase-Z1 | 69 | 17397 | 7.77 | 14 |
| gi|18859071* | Nucleoside diphosphate kinase 3 | 69 | 19558 | 7.68 | 7 |
| gi|126632622* | Novel protein containing a galactose binding Lectin domain | 67 | 19245 | 9.33 | 13 |
| gi|66773080 * | Mitochondrial ATP synthase beta subunit-like | 66 | 55080 | 5.25 | 4 |
| gi|38541767* | Ppia protein | 60 | 19745 | 9.30 | 13 |
| gi|1865782 | HSC70 protein | 58 | 71473 | 5.18 | 2 |
| gi|28279108 | Heat shock protein 8 | 58 | 71382 | 5.32 | 4 |
| gi|41152402* | Histone H2B 3 | 49 | 13940 | 10.31 | 11 |
| gi|41393113* | Collagen, type I, alpha 1b precursor | 46 | 137815 | 5.39 | 4 |
| gi|94732492 * | Ras homolog gene family, member F | 46 | 24035 | 9.00 | 6 |
| gi|47778620 * | Tryptophan hydroxylase D2 | 45 | 55686 | 6.56 | 1 |
| gi|68448517 * | Zona pellucida glycoprotein 3.2 precursor | 44 | 47365 | 4.92 | 2 |
| gi|326677766 * | PREDICTED: RIMS-binding protein 2-like | 41 | 138659 | 5.86 | 0 |
| gi|112419298 | Vtg3 protein | 40 | 60622 | 6.32 | 2 |
| gi|54400406 * | Glutaredoxin 3 | 39 | 36541 | 5.18 | 11 |
| gi|41152400* | Peptidylprolyl isomerase A, like | 37 | 17763 | 8.26 | 7 |
Discussion and Conclusion
The use of stem cells differentiation factors in anticancer therapy has enabled one of us to build up a model of cancer corresponding to reality [41]. Such a model, conceived in 2002, describes cancer as a consequence of two different processes, i) a process of maturation arrest of stem cells (hierarchical model) and ii) a process of deterministic chaos in which genetic and epigenetic alterations conduce a normal differentiated cell to be malignant (stochastic model). In fact, these two processes are not mutually exclusive and both have been described [42, 43].
Therefore, from this point of view, cancer cells can be defined as “cancer stem-like cells”, that according to their degree of malignancy, are considered blocked at a different phase of development. In fact, in tumors with an elevated degree of malignancy, such as acute lymphoblastic and myeloid leukemia, multipotent stem-like cells are present, whereas in tumors with lower malignancy, such as chronic lymphocytic leukemia, cells not yet completely differentiated are present, but towards a final differentiation.
In addition, cancer and stem cells share several characteristics. Firstly, they present oncofetal antigens, maintained during the phylogenesis [44] and specific receptor on the cellular membrane on which the stem cells differentiation factors probably act. It has already been mentioned above that such factors could activate pathways of cellular differentiation, that lead the cells to differentiate or to die, as usually occurs in the embryo (the apoptotic events in the embryo are many).
Furthermore, cancer and embryonic cells share common metabolic pathways such as APC/beta catenin/ TCF/Wnt and the Hedgehog/Smoothened/Patched pathways.
The gene configuration and the metabolism of cancer cells is actually very similar to that of stem cells: they both have active proto-oncogene and produce embryonic growth factors, present oncofetal antigens and they work with an aerobic metabolism.
Nevertheless, cancer cells and stem cells show an important difference. The problem of cancer cells is double: they present genetic mutations that are at the origin of malignancy and, at the same time, they show an imbalance of the epigenetic code. In contrast with normal stem cells, tumor cells are not able to complete their development and to differentiate because they lost informations, i.e. they experienced a mutation or epigenetic alterations in their code. The regulation of DNA informations using epigenetic regulators such as SCDSFs, taken in the intermediate-late stages of development of the embryo, transforms the cancer cells into normal cells or causes their apoptosis.
It is now emerging more and more clearly that the transcription factors, the microRNAs, the translational- and post-translational factors, play a fundamental role in the regulation of DNA informations and in regulating the cell life. In other words, the epigenetic regulators contained in SCDSFs are able to differentiate and regulate normal stem cells and cancer stem cells, deactivating genes that lead cancer stem cells to proliferate while activating new differentiating pathways.
Our studies have recently been confirmed by other experimental researches performed by some colleagues of the Children Hospital of Chicago [12]. In particular, they have confirmed that malignant melanoma reverts to a normal phenotype when it is in the environment of Zebrafish embryo.
Other extensive series of research confirm that tumors represent a problem of developmental biology. First of all, regarding the theories on attractors, it is to remember that more than 50 years ago, some authors [45] speculated that cancer could be represented as an escape from a fields like those which guide the embryonic development, hypothesis recently confirmed [46]. A similar hypothesis has been recently put forward again by other authors [47] who suggest that any change that conducts stem cells to evade from the its own niche would result in tumor formation. In addition it has already been demonstrated that tumor cells placed into a microenvironment, like that of an embryo, can change the malignant phenotype and reverse into normal cells [48, 49]. These data suggest that cancer can be considered an emergent property of living tissues under chronic stimulations, involving not merely DNA or single-somatic cell functions, but instead system features [50]. On the other hand, if we move from genomic alterations to cancer transcriptome, considered as “a whole”, as suggested by some authors, a more interesting picture emerges, from which it is possible observe a very great degree of order. In this vision tumors can be classified into a small number of discretely distinct groups, recording the organization of transcriptomes of cell types into groups of related tissues [51-54]. There are many studies that highlight the link between tumor malignancy and the presence of cancer stem cells [55] that seem to be resistant to conventional therapy, such as chemo- and radiotherapy. In the last 6-7 years scientific works in this field are so numerous that it is almost impossible to name all of them. Here we mention only those researches that demonstrated the presence of tumoral stem cell in breast cancer [56-61], lung cancer [62-65], prostate [66-68], ovary cancer [69-73], liver cancer [74-79], stomach cancer [80-84], colon cancer [85, 86, 87], pancreas cancer [88, 89, 90], glioblastoma multiforme [91-93], head and neck cancer [94-97]. On the other hand, it is known that malignancy of many haematological tumoral diseases is due to the presence of stem cells.
Regarding the interpretation of the results obtained by using SCDSFs for the prevention of the neurodegeneration and for the treatment of psoriasis, we can assume that: the differentiation factors are epigenetic regulators, that, on the one hand prevent the processes and the development of degenerative phenomena and, on the other hand, regulate the processes of abnormal cellular multiplication, as it comes for instance, in psoriasis, where the multiplication of cells of the epithelial basal layer is five-ten fold higher of that considered physiological. In this case we have demonstrated that the differentiation factors reduce the proliferation of the epidermal layers by normalizing it. In addition other researches demonstrated that it is possible to tune in fine way the fate of normal stem cells, like human mesenchimal stem cells using SCDSFs. In fact, if we use in specific way the different networks of substances present in the different stages of cell differentiation we can address stem cells toward the senescence or the apoptosis (late stages of differentiation) or, at the contrary, enhance stem cell expression of multipotency by activating both telomerase-dependent and -independent antagonists of cell senescence (early stage of differentiation). Noteworthy, different modulating effects can be obtained only with a specific network of SCDSFs. From this point of view, the experiments about the prevention of neurodegeneration are enlightening. In fact, to prevent neurodegeneration first of all we have to enhance stem cell expression of multipotency and then, we have to address stem cells toward the differentiation in neural cells. For these reasons all the different stage factors expressed during cell differentiation have to be used: only the redundancy of these factors could led to obtain significant results. These results make us to consider a major shift in scientific paradigm (from reductionism to complexity) for preparing new treatments for chronic and degenerative diseases. In fact, these diseases entail unexpected degree of complexity and disregulation, making the single-molecule-to-specific-target paradigm totally obsolete and inadequate. Rather, only a systemic approach can be envisioned as a successful strategy to deal with such complexity. We believe that time is ready for a “transdisciplinary approach” in the treatment of degenerative diseases involving multiple tissues and organs, to help users in a new culture of collaboration from different scientific disciplines joining together to combine their knowledge and come up with innovations, new therapeutic approaches and most of all the development of novel paradigms. This new culture is overdue to provide a reliable effort to help elderly people and anyone who suffers for degenerative diseases or cancer.
List of Abbreviations
- aa
Amino-acid
- Caco-2
Human Colon cancer cells
- DPBS
Dulbecco Phosphate Buffered Saline
- EGF
Epidermal Growth Factor
- FCS
Fetal Calf Serum
- 5-Fu
5-Fluorouracil
- hASCs
Human Adipose-derived Stem Cells
- hcc
Hepatocellular carcinoma
- HGF
Hepatocyte Growth Factor
- HSP
Heat Shock Protein
- iPS
Induced Pluripotent Stem cells
- LC-MS/MS
Liquid Chromatography–Mass Spectrometry
- MAPK
Mitogen-Activated Protein Kinase
- NMDA
N-Methyl-D-Aspartate
- PI
Propidium Iodide
- pI
Isoelectric point
- pRb
Retinoblastoma protein
- SCDSFs
Stem Cell Differentiation Stage Factors
- SDS-PAGE
Sodium Dodecyl Sulphate - PolyAcrylamide Gel Electrophoresis
REFERENCES
- 1.Einhorn L. Are there factors preventing cancer development during embryonic life? Oncodev. Biol. Med. 1983;4(3):219–229. [PubMed] [Google Scholar]
- 2.Lakshmi M.S., Sherbet G.V. In: Embryonic and Tumour Cell Interactions. Sherbet G.V., editor. New York: Karger Basel; 1974. pp. 380–399. [Google Scholar]
- 3.Brent R.L. Radiation teratogenesis. Teratology. 1980;21(3):281–298. doi: 10.1002/tera.1420210304. [DOI] [PubMed] [Google Scholar]
- 4.Pierce G.B. The cancer cell and its control by the embryo. Rous-Whipple Award lecture. Am. J. Pathol. 1983;113(1):117–124. [PMC free article] [PubMed] [Google Scholar]
- 5.Yu C.L., Tsai M.H. Fetal fetuin selectively induces apoptosis in cancer cell lines and shows anti-cancer activity in tumor animal models. Cancer Lett. 2001;166(2):173–184. doi: 10.1016/s0304-3835(01)00417-7. [DOI] [PubMed] [Google Scholar]
- 6.Papaioannou V.E., McBurney M.V., Gardner R.L., Evans M.J. Fate of teratocarcinoma cells injected into early mouse embryos. Nature. 1975;258(5530):70–73. doi: 10.1038/258070a0. [DOI] [PubMed] [Google Scholar]
- 7.Topczewska J.M., Postovit L.M., Margaryan N.V., Sam A., Hess A.R., Wheaton W.W., Nickoloff B.J., Topczewski J., Hendrix M.J. Embryonic and tumorigenic pathways converge via Nodal signaling: role in melanoma aggressiveness. Nat. Med. 2006;12(8):925–932. doi: 10.1038/nm1448. [DOI] [PubMed] [Google Scholar]
- 8.Kulesa P.M., Kasermeier-Kulesa J.C., Teddy J.M., Margaryan N.V., Seftor E.A., Seftor R.E., Hendrix M.J. Reprogramming metastatic melanoma cells to assume a neural crest cell-like phenotype in an embryonic microenvironment. Proc. Natl. Acad. Sci. USA. 2006;103(10):3752–3757. doi: 10.1073/pnas.0506977103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Webb C.G., Gootwine E., Sachs L. Developmental potential of myeloid leukemia cells injected into midgestation embryos. Dev. Biol. 1984;101(1):221–224. doi: 10.1016/0012-1606(84)90132-5. [DOI] [PubMed] [Google Scholar]
- 10.Weaver V.M., Petersen O.W., Wang F., Larabell C.A., Briand P., Damsky C., Bissell M.J. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin bloking antibodies. J. Cell Biol. 1997;137(1):231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coleman W.B., Wennerberg A.E., Smith G.J., Grisham J.W. Regulation of the differentiation of diploid and some aneuploid rat liver epithelial (stemlike) cells by the hepatic microenvironment. Am. J. Pathol. 1993;142(5):1373–1382. [PMC free article] [PubMed] [Google Scholar]
- 12.Postovit L.M., Maragaryan N.V., Seftor E.A., Kirschmann D.A., Lipavsky A., Wheaton W.W., Abbott D.E., Seftor R.E., Hendrix M.J. Human embryonic stem cell microenvironment suppresses the tumorigenic phenotype of aggressive cancer cells. Proc. Natl. Acad. Sci. USA. 2008;105(11):4329–4334. doi: 10.1073/pnas.0800467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallagher J.M., Komati H., Roy E., Nemer M., Latinkic B.V. Dissociation of cardiogenic and postnatal myocardial activities of GATA4. Mol. Cell. Biol. 2012;32(12):2214–2223. doi: 10.1128/MCB.00218-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rizzi R., Di Pasquale E., Portararo P., Papait R., Cattaneo P., Latronico M.V., Altomare C., Sala L., Zaza A., Hirsch E., Naldini L., Condorelli G., Bearzi C. Post-natal cardiomyocytes can generate iPS cells with an enhanced capacity toward cardiomyogenic re-differentation. Cell Death Differ. 2012;19(7):1162–1174. doi: 10.1038/cdd.2011.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ming G.L., Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70(4):687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livraghi T., Meloni F., Frosi A., Lazzaroni S., Bizzarri T.M., Frati L., Biava P.M. Treatment with stem cell differentiation stage factors of intermediate-advanced hepatocellular carcinoma: An open randomized clinical trial. Oncol. Res. 2005;15(7-8):399–408. doi: 10.3727/096504005776449716. [DOI] [PubMed] [Google Scholar]
- 17.Livraghi T., Ceriani R., Palmisano A., Pedicini V., Pich M.G., Tommasini M.A., Torzilli G. Complete response in 5 out of 38 patients with advanced hepatocellular carcinoma treated with stem cell differentiation stage factors: case reports from a single centre. Curr. Pharm. Biotechnol. 2011;12(2):254–260. doi: 10.2174/138920111794295855. [DOI] [PubMed] [Google Scholar]
- 18.Canaider S., Maioli M., Facchin F., Bianconi E., Santaniello S., Pigliaru G., Ljungberg L., Burigana F, Bianchi F., Olivi E., Tremolada C., Biava P.M., Ventura C. Human stem cell exposure to developmental stage Zebrafish extracts: A novel strategy for tuning stemness and senescence patterning. . Cell R4. 2014;2(5) [Google Scholar]
- 19.Di Pierro F., Negri M., Bollero C. Terapia della psoriasi. Efficacia clinica di un preparato multicomponente. Cosmetic Technol. 2009;12(2):13–17. [Google Scholar]
- 20.Harak H., Frosi A., Biava P.M. Studio clinico sull’efficacia e tollerabilita’ di una crema per uso topico nel trattamento della psoriasi. Med. Biol. 2012;3:27–31. [Google Scholar]
- 21.Calzavara-Pinton P., Rossi M. A topical remedy in association with phototherapy. Efficacy evaluation in patients suffering from moderate psoriasis. Hi. Tech. Dermo. 2012;1:41–47. [Google Scholar]
- 22.Plikus M.V., Van Spyk E.N., Pham K., Geyfman M., Kumar V., Takahashi J.S., Andersen B. The circadian clock in skin: Implications for adult stem cells, tissue regeneration, cancer, aging, and immunity. J. Biol. Rhythms. 2015:0748730414563537. doi: 10.1177/0748730414563537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou R., Liu R., Niu X., Chang W., Yan X., Wang C., Li J., An P., Li X., Yin G., Zhang K. Biological characteristics and gene expression pattern of bone marrow mesenchymal stem cells in patients with psoriasis. Exp. Dermatol. 2014;23(7):521–523. doi: 10.1111/exd.12446. [DOI] [PubMed] [Google Scholar]
- 24.Biava P.M., Bonsignorio D., Hoxa M. Cell proliferation curves of different human tumor lines after in vitro treatment with Zebrafish embryonic extracts. J. Tumor Marker Oncol. 2001;16(3):195–202. [Google Scholar]
- 25.Biava P.M., Carluccio A. Activation of anti-oncogene p53 produced by embryonic extracts in vitro tumor cells. J. Tumor Marker Oncol. 1977;12(4):9–15. [Google Scholar]
- 26.Biava P.M., Bonsignorio D., Hoxa M., Impagliazzo M., Facco R., Ielapi T., Frati L., Bizzarri M. Post-translational modification of the retinoblastoma protein (pRb) induced by in vitro administration of Zebrafish embryonic extracts on human kidney adenocarcinoma cell line. J. Tumor Marker Oncol. 2002;17(2):59–64. [Google Scholar]
- 27.Cucina A., Biava P.M., D’Anselmi F., Coluccia P., Conti F., di Clemente R., Miccheli A., Frati L., Gulino A., Bizzarri M. Zebrafish embryo proteins induce apoptosis in human colon cancer cells (Caco2). Apoptosis. 2006;11(9):1617–1628. doi: 10.1007/s10495-006-8895-4. [DOI] [PubMed] [Google Scholar]
- 28.D’Anselmi F., Cucina A., Biava P.M., Proietti S., Coluccia P., Frati L., Bizzarri M. Zebrafish stem cell differentiation stage factors suppress Bcl-xL release and enhance 5-Fu-mediated apoptosis in colon cancer cells. Curr. Pharm. Biotechnol. 2011;12(2):261–267. doi: 10.2174/138920111794295864. [DOI] [PubMed] [Google Scholar]
- 29.Biava P.M., Nicolini A., Ferrari P., Carpi A., Sell S. A systemic approach to cancer treatment: tumor cell reprogramming focused on endocrine-related cancers. Curr. Med. Chem. 2014;21(9):1072–1081. doi: 10.2174/0929867321666131201143124. [DOI] [PubMed] [Google Scholar]
- 30.Bianchi F., Maioli M., Leonardi E., Olivi E., Pasquinelli G., Valente S., Mendez A.J., Ricordi C., Raffaini M., Tremolada C., Ventura C. A new non enzymatic method and device to obtain a fat tissue derivative high enriched in pericyte-like elements by mild mechanical forces from human kipoaspirates. Cell Transplant. 2013;22(11):2063–2077. doi: 10.3727/096368912X657855. [DOI] [PubMed] [Google Scholar]
- 31.Conner E.A., Teramoto T., Wirth P.J., Kiss A., Garfield S., Thorgeirsson S.S. HGF-mediated apoptosis via p53/bax-independent pathway activating JNK1. Carcinogenesis. 1999;20(4):583–590. doi: 10.1093/carcin/20.4.583. [DOI] [PubMed] [Google Scholar]
- 32.Feng R., Zhou S., Liu Y., Song D., Luan Z., Dai X., Li Y., Tang N., Wen J., Li L. Sox2 protects neural stem cells from apoptosis via up regulating survivin expression. Biochem. J. 2013;450(3):459–468. doi: 10.1042/BJ20120924. [DOI] [PubMed] [Google Scholar]
- 33.Park I.K., Qian D., Kiel M., Becker M.W., Pihalja M., Weissman I.L., Morrison S.J., Clarke M.F. Bmi-1 is required for maintenance of adult self-renewing hematopioetic stem cells. Nature. 2003;423(6937):302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 34.Gardoni F., Bellone C., Viviani B., Marinovich M., Meli E., Pellegrini-Giampietro D.E., Cattabeni F., Di Luca M. Lack of PSD-95 drives hippocampal neuronal cell death through activation of an alpha CaMKII transduction pathway. Eur. J. Neurosci. 2002;16(5):777–786. doi: 10.1046/j.1460-9568.2002.02141.x. [DOI] [PubMed] [Google Scholar]
- 35.Pellegrini-Giampietro D.E., Cozzi A., Peruginelli F., Leonardi P., Meli E., Pellicciari R., Moroni F. 1-Aminoindan-1,5-dicarboxylic acid and (S)-(+)-2-(3’-carboxybicyclo[1.1.1] pentyl)-glycine, two mGlu1 receptor-preferring antagonists, reduce neuronal death in in vitro and in vivo models of cerebral ischaemia. Eur. J. Neurosci. 1999;11(10):3637–3647. doi: 10.1046/j.1460-9568.1999.00786.x. [DOI] [PubMed] [Google Scholar]
- 36.Norata G.D., Biava P.M., Di Pierro F. The Zebrafish embryo derivative affects cell viability of epidermal cells: a possible role in the treatment of psoriasis. G. Ital. Dermatol. Venereol. 2013;148(5):479–483. [PubMed] [Google Scholar]
- 37.Shi Q., Jackowski G. New York: Oxford University Press Inc:; 1998. Gel Electrophoresis of Proteins: A Practical Approach ; pp. 13–29. [Google Scholar]
- 38.Kang D.H., Gho Y.S., Suh M.K., Kang C.H. Highly sensitive and fast protein detection with coomassie brilliant blue in sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Bull. Korean Chem. Soc. 2002;23:1511–1512. [Google Scholar]
- 39.Della Corte A., Tamburrelli C., Crescente M., Giordano L., D'Imperio M., Di Michele M., Donati M.B., De Gaetano G., Rotilio D., Cerletti C. Platelet proteome in healthy volunteers who smoke. Platelets. 2012;23(2):91–105. doi: 10.3109/09537104.2011.587916. [DOI] [PubMed] [Google Scholar]
- 40.Lucitt M.B., Price T.S., Pizarro A., Wu W., Yocum A.K., Seiler C., Pack M.A., Blair I.A., Fitzgerald G.A., Grosser T. Analysis of the Zebrafish proteome during embryonic development. Mol. Cell. Proteomics. 2008;7(5):981–994. doi: 10.1074/mcp.M700382-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biava P.M., Bonsignorio D. Cancer and cell differentiation: a model to explain malignancy. J. Tumor Marker Oncol. 2002;17(2):47–54. [Google Scholar]
- 42.Shackleton M., Quintana E., Fearon E.R., Morrison S.J. Heterogeneity in cancer: Cancer stem cells versus clonal evolution. Cell. 2009;138(5):822–829. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 43.Visvader J.E., Lindeman G.J. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10(6):717–728. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 44.Biava P.M., Monguzzi A., Bonsignorio D., Frosi A., Sell S., Klavins J.V. Xenopus laevis Embryos share antigens with Zebrafish Embryos and with human malignant neoplasms. J. Tumor Marker Oncol. 2001;16(3):203–206. [Google Scholar]
- 45.Waddington C.H. Cancer and theory of organizers. Nature. 1935;135:606–608. [Google Scholar]
- 46.Bizzarri M., Cucina A., Biava P.M., Proietti S., D'Anselmi F., Dinicola S., Pasqualeto A., Lisi E. Embryonic morphogenetic field induce phenotipic reversion in cancer cells. Curr. Pharm. Biotechnol. 2011;12:243–253. doi: 10.2174/138920111794295701. [DOI] [PubMed] [Google Scholar]
- 47.Ruiz-Vela A., Aguillar-Gallardo C., Simon C. Building a framework for embryonic microenvironments and cancer stem cells. Stem Cell Rev. 2009;5:319–327. doi: 10.1007/s12015-009-9096-7. [DOI] [PubMed] [Google Scholar]
- 48.Kenny P.A., Bissel M.J. Tumor reversion: correction of malignant behaviour by microenvironmental cues. Int. J. Cancer. 2003;107:688–695. doi: 10.1002/ijc.11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diez-Torre A., Andrade R., Eguizabal C., Lopez E., Arluzea J., Siliò M., Aréchaga L. Reprogramming of melanoma cells by embryonic microenvironment. Int. Dev. Biol. 2009;53:1563–1568. doi: 10.1387/ijdb.093021ad. [DOI] [PubMed] [Google Scholar]
- 50.Huang S., Ernberg I., Kauffman S. Cancer attractors. A system view of tumors from a gene network dynamics and developmental perspectives. Semin. Cell Dev. Biol. 2009;20:869–876. doi: 10.1016/j.semcdb.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo Y., Eichler G.S., Feng Y., Ingberg D.E., Huang S. Towards a holistic; yet gene-centered analysis of gene expression profiles: a case study of human lung cancers. J. Biomed. Biotechnol. 2006:69141. doi: 10.1155/JBB/2006/69141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su A.L., Wiltshire T., Batalo S., Lapp H., Ching K.A., Block D.A. Gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sonnenschein C., Davis B., Soto A.M. A novel pathogenic classification of cancers. Cancer Cell Int. 2014;14(1):113. doi: 10.1186/s12935-014-0113-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bizzarri M., Cucina A. Tumor and the microenvironment: a chance to reframe the paradigm of carcinogenesis? 2014. [DOI] [PMC free article] [PubMed]
- 55.Reya T., Morrison S.J., Clarke M.F., Weissman I.L. Stem cells, cancer and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 56.Chen J., Chen Z.L. Technology update for the sorting and identification of breast cancer stem cells. Chin. J. Cancer. 2010;29(3):265–269. doi: 10.5732/cjc.009.10642. [DOI] [PubMed] [Google Scholar]
- 57.Roesler R., Cornelio D.B., Abujamra A.L., Schwartsmann G. HER2 as a cancer stem-cell target. Lancet Oncol. 2010;11(3):225–226. doi: 10.1016/S1470-2045(09)70404-8. [DOI] [PubMed] [Google Scholar]
- 58.Wu W. Patents related to cancer stem cell research. Recent Pat. DNA Gene Seq. 2010;4(1):40–45. doi: 10.2174/187221510790410840. [DOI] [PubMed] [Google Scholar]
- 59.Park S.Y., Lee H.E., Li H., Shipitsin M., Gelman R., Polyak K. Heterogeneity for stem cell-related markers according to tumor subtype and histologic stage in breast cancer. Clin. Cancer Res. 2010;16(3):876–887. doi: 10.1158/1078-0432.CCR-09-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lawson J.C., Blatch G.L., Edkins A.L. Cancer stem cells in breast cancer and metastasis. Breast Cancer Res. Treat. 2009;118(2):241–254. doi: 10.1007/s10549-009-0524-9. [DOI] [PubMed] [Google Scholar]
- 61.Luo J., Yin X., Ma T., Lu J. Stem cells in normal mammary gland and breast cancer. Am. J. Med. Sci. 2010;339(4):366–370. doi: 10.1097/MAJ.0b013e3181cad964. [DOI] [PubMed] [Google Scholar]
- 62.Spiro S.G., Tanner N.T., Silvestri G.A., Janes S.M., Lim E., Vansteenkiste J.F., Pirker R. Lung cancer: progress in diagnosis staging and therapy. Respirology. 2010;15(1):44–50. doi: 10.1111/j.1440-1843.2009.01674.x. [DOI] [PubMed] [Google Scholar]
- 63.Gorelik E., Lokshin A., Levina V. Lung cancer stem cells as a target for therapy. Anticancer. Agents Med. Chem. 2010;10(2):164–171. doi: 10.2174/187152010790909308. [DOI] [PubMed] [Google Scholar]
- 64.Sullivan J.P., Minna J.D., Shay J.W. Evidence for self-renewing lung cancer stem cells and their implications in tumor initiation, progression and targeted therapy. Cancer Metastasis Rev. 2010;29(1):61–72. doi: 10.1007/s10555-010-9216-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Westhoff B., Colaluca I.N., D’Ario G., Donzelli M., Tosoni D., Volorio S., Pelosi G., Spaggiari L., Mazzarol G., Viale G., Pece S., Di Fiore P.P. Alterations of the Notch pathway in lung cancer. Proc. Natl. Acad. Sci. USA. 2009;106(52):22293–22298. doi: 10.1073/pnas.0907781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lawson D.A., Zong Y., Memarzadeh S., Xin L., Huang J., Witte O.N. Basal epithelial stem cells are efficient targets for prostate cancer initiation. Proc. Natl. Acad. Sci. USA. 2010;107(6):2610–2615. doi: 10.1073/pnas.0913873107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lang S.H., Anderson E., Fordham R., Collins A.T. Modeling the prostate stem cell niche: an evaluation of stem cell survival and expansion in vitro. Stem Cells Dev. 2010;19(4):537–546. doi: 10.1089/scd.2009.0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Joung J.Y., Cho K.S., Kim J.E., Seo H.K., Chung J., Park W.S., Choi M.K., Lee K.H. Prostate stem cell antigen mRNA in peripheral blood as a potential predictor of biochemical recurrence of metastatic prostate cancer. J. Surg. Oncol. 2010;101(2):145–148. doi: 10.1002/jso.21445. [DOI] [PubMed] [Google Scholar]
- 69.Liu T., Cheng W., Lai D., Huang Y., Guo L. Characterization of primary ovarian cancer cells in different culture systems. Oncol. Rep. 2010;23(5):1277–1284. doi: 10.3892/or_00000761. [DOI] [PubMed] [Google Scholar]
- 70.Fong M.Y., Kakar S.S. The role of cancer stem cells and the side population in epithelial ovarian cancer. Histol. Histopathol. 2010;25(1):113–120. doi: 10.14670/HH-25.113. [DOI] [PubMed] [Google Scholar]
- 71.Murphy S.K. Targeting ovarian cancer-initiating cells. Anticancer. Agents Med. Chem. 2010;10(2):157–163. doi: 10.2174/187152010790909272. [DOI] [PubMed] [Google Scholar]
- 72.Peng S., Maihle N.J., Huang Y. Pluripotency factors Lin 28 and Oct 4 identify a sub-population of stem cell-like cells in ovarian cancer. Oncogene. 2010;29(14):2153–2159. doi: 10.1038/onc.2009.500. [DOI] [PubMed] [Google Scholar]
- 73.Kusumbe A.P., Bapat S.A. Cancer stem cells and aneuploid populations within developing tumors are the major determinants of tumor dormancy. Cancer Res. 2009;69(24):9245–9253. doi: 10.1158/0008-5472.CAN-09-2802. [DOI] [PubMed] [Google Scholar]
- 74.Tomuleasa C., Soritau O., Rus-Ciuca D., Pop T., Todea D., Mosteanu O., Pintea B., Foris V., Susman S., Kacso G., Irimie A. Isolation and characterization of hepatic cells with stem-like properties from hepatocellular carcinoma. J. Gastrointestin. Liver Dis. 2010;19(1):61–67. [PubMed] [Google Scholar]
- 75.Zou G.M. Liver cancer stem cells as an important target in liver cancer therapies. Anticancer. Agents Med. Chem. 2010;10(2):172–175. doi: 10.2174/187152010790909263. [DOI] [PubMed] [Google Scholar]
- 76.Lee T.K., Castilho A., Ma S., Ng I.O. Liver cancer stem cells: implications for new therapeutic target. Liver Int. 2009;29(7):955–965. doi: 10.1111/j.1478-3231.2009.02040.x. [DOI] [PubMed] [Google Scholar]
- 77.Marquardt J.U., Thorgeirsson S.S. Stem Cells in hepatocarcinogenesis: Evidence from genomic data. Semin. Liver Dis. 2010;30(1):26–34. doi: 10.1055/s-0030-1247130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kung J.W., Currie I.S., Forbes S.J., Ross J.A. Liver development, regeneration, and carcinogenesis. 2010. [DOI] [PMC free article] [PubMed]
- 79.Gai H., Nguyen D.M., Moon Y.J., Aguila J.R., Fink L.M., Ward D.C., Ma Y. Generation of murine hepatic lineage cells from induced pluripotent stem cells. Differentiation. 2010;79(3):171–181. doi: 10.1016/j.diff.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 80.Correia M., Machado J.C., Ristimaki A. Basic aspects of gastric cancer. Helicobacter. 2009;14(1):36–40. doi: 10.1111/j.1523-5378.2009.00696.x. [DOI] [PubMed] [Google Scholar]
- 81.Takaishi S., Okumura T., Tu S., Wang S.S., Shibata W., Vigneshwaran R., Gordon S.A., Shimada Y., Wang T.C. Identification of gastric cancer stem cells using the surface marker CD44. Stem Cells. 2009;27(5):1006–1020. doi: 10.1002/stem.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nishii T., Yashiro M., Shinto O., Sawada T., Ohira M., Hirakawa K. Cancer stem cell-like SP cells have a high adhesion ability to the peritoneum in gastric carcinoma. Cancer Sci. 2009;100(8):1397–1402. doi: 10.1111/j.1349-7006.2009.01211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen Z., Xu W.R., Qian H., Zhu W., Bu X.F., Wang S., Yan Y.M., Mao F., Gu H.B., Cao H.L., Xu X.J. Oct4, a novel marker for human gastric cancer. J. Surg. Oncol. 2009;99(7):414–419. doi: 10.1002/jso.21270. [DOI] [PubMed] [Google Scholar]
- 84.Kang D.H., Han M.E., Song M.H., Lee Y.S., Kim E.H., Kim H.J., Kim G.H., Kim D.H., Yoon S., Baek S.Y., Kim B.S., Kim J.B., Oh S.O. The role of hedgehog signaling during gastric regeneration. J. Gastroenterol. 2009;44(5):372–379. doi: 10.1007/s00535-009-0006-1. [DOI] [PubMed] [Google Scholar]
- 85.Yeung T.M., Ghandhi S.C., Wilding J.L., Muschel R., Bodmer W.F. Cancer stem cells from colorectal cancer derived cell lines. Proc. Natl. Acad. Sci. USA. 2010;107(8):3722–3727. doi: 10.1073/pnas.0915135107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gulino A., Ferretti E., De Smaele E. Hedgehog signaling in colon cancer and stem cells. EMBO Mol. Med. 2009;1(6-7):300–302. doi: 10.1002/emmm.200900042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thenappan A., Li Y., Shetty K., Johnson L., Reddy E.P., Mishra L. New therapeutic targeting colon cancer stem cells. Curr. Colorectal Cancer Rep. 2009;5(4):209. doi: 10.1007/s11888-009-0029-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rasheed Z.A., Yang J., Wang Q., Kowalski J., Freed I., Murter C., Hong. S.M., Koorstra J.B., Rajeshkumar N.V., He X., Goggins M., Iacobuzio-Donahue C., Berman D.M., Laheru D., Jimeno A., Hidalgo M., Maitra A., Matsui W. Prognostic significance of tumorigenic cells with mesenchymal features in pancreatic adenocarcinoma. J. Natl. Cancer Inst. 2010;102(5):340–351. doi: 10.1093/jnci/djp535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Puri S., Hebrok M. Cellular plasticity within pancreas-lessons learned from development. Dev. Cell. 2010;18(3):342–356. doi: 10.1016/j.devcel.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Quante M., Wang T.C. Stem cells in gastroenterology and hepatology. Nat. Rev. Gastroenterol. Hepatol. 2009;6(12):724–737. doi: 10.1038/nrgastro.2009.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sato A., Sakurada K., Kumabe T., Sasajima T., Beppu T., Asano K., Ohkuma H., Ogawa A., Mizoi K., Tominaga T., Kitanaka C., Kayama T., Tohoku Brain Tumor Study Group Association of stem cell marker CD133 expression with dissemination of glioblastoma. Neurosurg. Rev. 2010;33(2):175–183. doi: 10.1007/s10143-010-0239-8. [DOI] [PubMed] [Google Scholar]
- 92.Di Tomaso T., Mazzoleni S., Wang E., Sovena G., Clavenna D., Franzin A., Mortini P., Ferrone S., Doglioni C., Marincola F.M., Galli R., Parmiani G., Maccalli C. Immunobiological characterization of cancer stem cells isolated from glioblastoma patients. Clin. Cancer Res. 2010;16(3):800–813. doi: 10.1158/1078-0432.CCR-09-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ji J., Black K.L., Yu J.S. Glioma stem cell research for the development of immunotherapy. Neurosurg. Clin. N. Am. 2010;21(1):159–166. doi: 10.1016/j.nec.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ailles L., Prince M. Cancer stem cells in head and neck squamous cell carcinoma. Methods Mol. Biol. 2009;568:175–193. doi: 10.1007/978-1-59745-280-9_11. [DOI] [PubMed] [Google Scholar]
- 95.Zhang P., Zhang Y., Mao L., Zhang Z., Chen W. Side population in oral squamous cell carcinoma possesses tumor stem cell phenotype. Cancer Lett. 2009;277(2):227–234. doi: 10.1016/j.canlet.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 96.Brunner M., Thurnher D., Heiduschka G., Grasl M.Ch., Brostjan C., Erovic B.M. Elevated levels of circulating endothelial progenitor cells in head and neck cancer patients. J. Surg. Oncol. 2008;98(7):545–550. doi: 10.1002/jso.21139. [DOI] [PubMed] [Google Scholar]
- 97.Zhang Q., Shi S., Yen Y., Brown J., Ta J.Q., Le A.D. A subpopulation of CD133(+) cancer stem-like cells characterized in human oral squamous cell carcinoma confer resistance to chemotherapy. Cancer Lett. 2010;289(2):151–160. doi: 10.1016/j.canlet.2009.08.010. [DOI] [PubMed] [Google Scholar]


