Abstract
Ohr and OsmC proteins comprise two subfamilies within a large group of proteins that display Cys-based, thiol dependent peroxidase activity. These proteins were previously thought to be restricted to prokaryotes, but we show here, using iterated sequence searches, that Ohr/OsmC homologs are also present in 217 species of eukaryotes with a massive presence in Fungi (186 species). Many of these eukaryotic Ohr proteins possess an N-terminal extension that is predicted to target them to mitochondria. We obtained recombinant proteins for four eukaryotic members of the Ohr/OsmC family and three of them displayed lipoyl peroxidase activity. Further functional and biochemical characterization of the Ohr homologs from the ascomycete fungus Mycosphaerella fijiensis Mf_1 (MfOhr), the causative agent of Black Sigatoka disease in banana plants, was pursued. Similarly to what has been observed for the bacterial proteins, we found that: (i) the peroxidase activity of MfOhr was supported by DTT or dihydrolipoamide (dithiols), but not by β-mercaptoethanol or GSH (monothiols), even in large excess; (ii) MfOhr displayed preference for organic hydroperoxides (CuOOH and tBOOH) over hydrogen peroxide; (iii) MfOhr presented extraordinary reactivity towards linoleic acid hydroperoxides (k=3.18 (±2.13)×108 M−1 s−1). Both Cys87 and Cys154 were essential to the peroxidase activity, since single mutants for each Cys residue presented no activity and no formation of intramolecular disulfide bond upon treatment with hydroperoxides. The pKa value of the Cysp residue was determined as 5.7±0.1 by a monobromobimane alkylation method. Therefore, eukaryotic Ohr peroxidases share several biochemical features with prokaryotic orthologues and are preferentially located in mitochondria.
Keywords: Ohr/OsmC, Thiol-dependent peroxidases, Phylogeny
Graphical abstract
Highlights
-
•
Ohr/OsmC proteins are also present in lower eukaryotic organisms.
-
•
While Ohr proteins are massively present among Fungi, OsmC proteins are restricted to the cellular slime molds.
-
•
Eukaryotic Ohr and OsmC present a thiol dependent peroxidase activity similar to the bacterial counterparts.
-
•
Most of these eukaryotic enzymes are predominantly present in mitochondria.
1. Introduction
Organic hydroperoxide resistance (Ohr) proteins are Cys-based, thiol dependent peroxidases that belong to a family of proteins called Ohr/OsmC. OsmC (Osmotically inducible protein) are structurally related to Ohr enzymes [1] and together define two subfamilies that have their peroxidase activities well characterized within Ohr/OsmC family [2], [3], [4]. Members of a third group remain poorly characterized [1].
The physiological role played by Ohr and OsmC has been linked to the defense against organic hydroperoxide insults [1], [5], [6], [7], [8], [9], [10], [11]. Ohr and OsmC are structurally distinct from peroxiredoxin (Prx) and glutathione-peroxidase (Gpx) enzymes [1], [2], although all are Cys-based, thiol dependent peroxidases. While Prx and Gpx enzymes are ubiquitously distributed in all domains of life, Ohr/OsmC proteins were thought to be present only in Archaea and Eubacteria [2], [6]. Contrary to Ohr enzymes, most Prx enzymes are highly reactive towards H2O2. One exception is Tpx from E. coli that similarly to Ohr enzymes also display higher specificity to organic peroxides over H2O2 [12], although the large majority of these peroxiredoxins are found in bacteria [13].
The catalytic mechanism of hydroperoxide reduction by Ohr and OsmC proteins is centered on a pair of redox-active cysteines, named peroxidatic (Cp) and resolving (Cr) cysteines, resembling that of the atypical 2-Cys Prxs. Ohr and OsmC are functionally dimeric and the cysteine residues are positioned in each monomer as part of two identical active sites located at opposing sides [2]. Two other residues of the active site also participate in catalysis: an arginine (Arg) and a glutamic acid (Glu) [3], [14]. The peroxidase cycle starts with the nucleophilic attack of Cysp towards the hydroperoxide. Upon hydroperoxide reduction to its corresponding alcohol, the Cp is oxidized to the sulfenic acid (Cys-SOH) intermediate, which readily reacts with Cr, giving rise to an intramolecular disulfide bond [14]. A new cycle begins when the disulfide bond is reduced back to the dithiolic form. Lipoyl groups covalently attached to some proteins are the biological reductants of these intramolecular disulfides [4]. Recently, we demonstrated that Ohr enzymes display high specificity for fatty acid hydroperoxides and peroxynitrite as oxidizing substrates [11].
Here, based on an in-depth sequence analysis, we describe the occurrence and distribution of Ohr and OsmC peroxidases in the Eukarya domain. OsmC proteins were only found in Dictyostelia, whereas Ohr members are predominantly present in Fungi (mainly Ascomycota and Basidiomycota). Four recombinant eukaryotic proteins from the Ohr/OsmC family were purified, three of which displayed thiol peroxidase activity. One of these, namely Ohr from the ascomycota fungus Mychosphaerella fijiensis Mf_1 (MfOhr), was further characterized, and its presence in the mitochondria of this fungus was demonstrated.
2. Material and methods
2.1. Dataset source and sequence extraction
The amino acid sequence from Xylella fastidiosa 9a5c strain was used as query for search against NCBI nr sequence database using delta-BLAST via NCBI website [15] (June of 2016) and the profile Hidden Markov Model (HMM) iterative method implemented in Jackhmmer 1.9 web server [16]. We conducted the search against NCBI nr sequence database using default options until convergence. The searches were restricted to the Eukarya Domain. Redundant entries and truncated sequences (less than 100 amino acids) were removed using CD-HIT software [17].
2.2. Primary sequence clustering
We identified members of the Ohr and OsmC subfamilies in Eukarya using sequence motifs previously described [1], [6]. Additional motifs were detected using alignments of eukaryotic Ohr sequences with structurally solved Ohr (PDB: 1ZB8, from X. fastidiosa; 1USP, from Deinococcus radiodurans; 3LUS, from Vibrio cholerae; 1N2F, from Pseudomonas aeruginosa) or OsmC (1NYE, from Escherichia coli) proteins. We curated the alignments manually, guided by successive multiple alignments runs generated by MAFFT operating with default sets [18]. The input sequences were collected by delta-blast and jackhmmer searches described in the previous section.
2.3. Phylogenetic analysis
Maximum Likelihood (ML) inference of phylogenetic trees was based on the manually curated MAFFT alignment and the RAxML software [19] and applied to all non-redundant sequences retrieved or only sequences from the Ohr subfamily. For inference, we used Whelan-Goldman (WAG) model of amino acid evolution with rate heterogeneity modeled by a GAMMA distribution and 1000 rapid bootstrap resampled estimates of log-likelihood (RELL bootstrap). The resulting phylogeny was prepared for visualization using Tree Editor from the MEGA 7 software [20].
2.4. Strains and growth conditions
E. coli strains were grown in Lysogenic Broth (LB) medium at 37 °C supplemented with ampicillin (100 μg/mL). Mycosphaerella fijiensis Mf_1 was grown in Potato Dextrose Medium (PDB) at room temperature supplemented with streptomycin (100 μg/mL) and chloramphenicol (100 μg/mL). Dictyostelium discoideum AX4 cells were grown axenically in liquid maltose HL-5 modified medium [21] supplemented with ampicillin (100 μg/mL) and streptomycin (300 μg/mL) at 22 °C.
2.5. Cloning procedures
To amplify ohr (MYCFIDRAFT_54770) and osmC (DDB_G0268884) genes without introns, samples of total RNA from germinated conidia of M. fijiensis Mf-1 and D. discoideum AX4 cells, respectively, were extracted using Trizol reagent (Ambion). RNA samples were treated with RNase-Free DNase I (Ambion) and submitted to reverse transcription (SuperScript II) using Oligo-dT to produce cDNA. To clone Mfohr into pET15b (Novagen®) and DdosmC into pPROEX expression vectors, sequences were amplified from appropriated cDNAs by PCR using the oligo pairs (5′→3′): Fow_TTAGCATATGGCTTCCGTAAGAGCATTC/Rev_TTAGGGATCCCGTCCCGCTCTATCCAATAA and Fow_AGTCATATGAGCATTAGTAATAAAATTATTGGATCAGC/Rev_AGTGGATCCCAAAAACAAATGGTGAGAAATCTG, respectively. The restriction sites for NdeI and BamHI are depicted by bold letters. Additionally, for Mfohr gene, a second PCR was performed using the same conditions described above using forward oligo (5′→3′) TTAGCATATGTCCCCGCCATTCTACACAGCCCAT, to produce a version of the protein MfOhr without the first 33 amino acid residues (MfOhrdel). The ohr gene from Fusarium oxysporum f. sp. cubense (Foohr) and Ohr-like (named as osmC gene by [22]) from Trichomonas vaginalis (TvosmC) were commercially synthesized by GenScript USA Inc., containing the sites for NheI and BamHI restriction enzymes in the flanking regions. The fragments that corresponded to the Foohr and Tvosmc genes were digested from pUC57 using NdeI and BamHI restriction enzymes and subcloned into pET15b. Fidelity of all sequences was checked by chain termination sequencing method using T7 promoter and terminator oligonucleotides.
2.6. Protein purification
Expression of recombinant MfOhr, MfOhrdel, FoOhr, DdOsmC or TvOsmC was induced by 0.1 mM of isopropyl 1-thio-β-D-galactopyranoside (IPTG) for 16 h at 20 °C in exponential culture (OD600 0.5) of E. coli BL21 (DE3) CodonPlus (Agilent) harboring the appropriate expression vectors with moderate shaking. Then, cells were harvested by centrifugation and resuspended in the lysis buffer (500 mM NaCl, 20 mM sodium phosphate pH 7.4, 0.2 mg/mL lysozyme, 1 mM PMSF and 20 mM imidazole). Cells were disrupted by sonication (ten alternating cycles of 15 s of sonication 30% amplitude and 1 min on ice bath). Cell debris were separated from the supernatant by centrifugation at 15.000 rpm at 4 °C during 40 min. The supernatant was filtered using a 0.45 µm pore membrane and all expressed proteins were affinity purified (Ni-NTA Agarose column, Qiagen) with a peristaltic pump. The charged resin was washed sequentially with 3 column volumes of washing buffer (500 mM NaCl, 20 mM sodium phosphate pH 7.4) containing 50 mM and 100 mM imidazole and eluted with 3 column volumes of elution buffer (500 mM NaCl, 20 mM sodium phosphate pH 7.4 and 500 mM imidazole). Buffer exchange and concentration of purified proteins were performed in an Amicon Centrifugal 10 MW device (Millipore®). Protein purity was checked by SDS-PAGE and protein concentration was spectrophotometrically determined by its absorbance at 280 nm (for MfOhr, εox =9970 and εred=10,095; for MfOhrdel, εox=4595 and εred=4470 M−1 cm−1; for FoOhr, εox=11,585 and εred=11,460 M−1 cm−1, for DdOsmC, εox=10,220 and εred=9970 and for TvOsmC, εox =15,720 and εred=15,470, according to ProtParam tool [23]).
2.7. Reduction of peroxidases with DTT
In some assays, Cys-based peroxidases (MfOhrdel or AhpE) were pre-reduced by 50 mM of DTT for 16 h at 4 °C, in the presence of 500 mM NaCl and 20 mM sodium phosphate pH 7.4. Excess of DTT was eliminated by two rounds of size-exclusion chromatography (HiTrap Dessalting, GE HealthCare) in a buffer (500 mM NaCl and 20 mM sodium phosphate pH 7.4) previously purged with N2. The efficiency of this procedure was ascertained by the DTNB method [24].
2.8. Thiol dependent peroxidase activity assays
Reductions of cumene hydroperoxide (CuOOH), tert-butyl hydroperoxide (tBOOH) or hydrogen peroxide (H2O2) were monitored by FOX assay [25] using DTT, dihydrolipoamide (DHLA), β-mercaptoethanol or glutathione as reductants.
Peroxidase activities were also analyzed by the lipoamide/lipoamide dehydrogenase coupled assay, following absorbance decay at 340 nm, as a consequence of NADH oxidation [4], [26], [27]. In this assay, the reaction was carried out at distinct concentrations of Ohr or OsmC enzymes as indicated in the legend of Fig. 4.
Fig. 4.
Thiol dependent peroxidase activity of eukaryotic Ohr and OsmC enzymes. Peroxidase activity assay of selected eukaryotic Ohr and OsmC proteins was assessed by the lipoamide/ lipoamide dehydrogenase coupled assay. The reactions were performed with 0.625 µM of MfOhr and FoOhr (A) or 5 µM of DdOsmC (B) in the presence of 50 µM of reduced lipoamide, 100 µM of DTPA, 0.5 µM of Lpd (recombinant Dihydrolipoamide dehydrogenase from X. fastidiosa) and 200 µM of NADH in 50 µM of sodium phosphate pH 7.4. Reactions were initiated by addition of 200 µM of CuOOH. As positive controls, we also performed the same assay using bacterial recombinant enzymes 0.1 µM PaOhr (A) or 5 µM of EcOsmC (B).
2.9. Site directed mutagenesis
The oligo pairs (5′→3′) used to mutated Cp to serine (C87S) were C87SF TACGGAGCTTCCTTCCAAG and C87SR CTTGGAAGGAAGCTCCGTA and Cr to serine (C154S) were C154SF AAGGAGGTCAGTCCGTATAGC and C154SR GCTATACGGACTGACCTCCTT, using the QuickChange II Site-Directed Mutagenesis Kit (Agilent Technologies). The bold letters indicate the mutate nucleotide(s).
2.10. Kinetics of linoleic acid hydroperoxide (LAOOH) reduction by MfOhrdel - AhpE competition assay
The rate constant for the reduction of LAOOH by MfOhr was calculated according to a competitive assay previously described [11] that takes advantage of the redox-dependent changes in the intrinsic fluorescence of AhpE, a Cys-based peroxidase from Mycobacterium tuberculosis (MtAhpE) [6]. Briefly, Ohr and AhpE were pre-reduced with 50 mM of DTT as described above. Stopped-flow fluorescence measurements were performed using a commercially available stopped-flow device (SFA-20 Rapid Kinetics Spectrometer Accessory, TgK Scientific, United Kingdom, UK) coupled to a Varian Cary Eclipse Fluorescence Spectrophotometer (Agilent Technologies).
2.11. pKa determination of CP residue from MfOhrdel WT
The pKa of the thiolate in Cp from MfOhrdel was determined using the monobromobimane (mBrB) alkylation method that generates a fluorescent product detected at λexc 396 nm and λem 482 nm [29]. MfOhrdel was pre-reduced by DTT as described above. The assays were performed in flat-bottom white polystyrene 96-well plates (Costar) in triplicates, using Varian Cary Eclipse fluorescence spectrophotometer, operating at medium voltage with both emission and excitation slit of 5 nm. Immediately after the end of the reaction, the pH of samples was checked. An additional blank reaction was performed in absence of thiols, to determine if other components in buffers might interfere with the reaction.
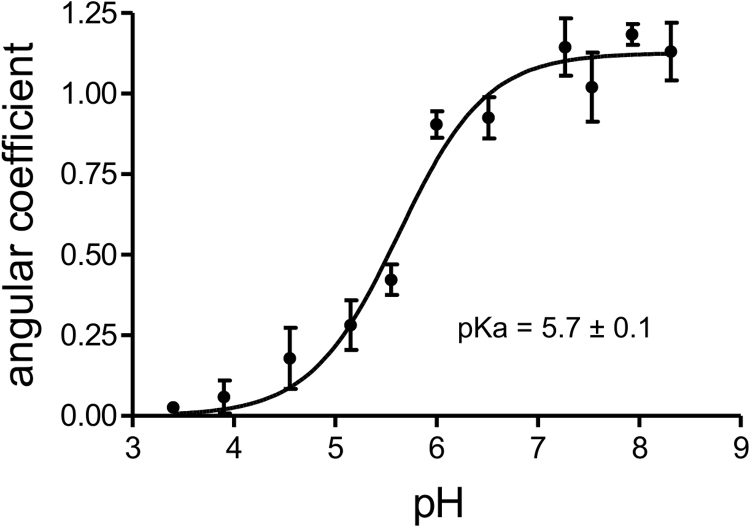
The angular coefficients were calculated using time points that included at least the initial 10 min of reaction that were fitted in a straight line. The curves displayed in Fig. 9 were obtained by non-linear regression using Henderson-Hasselbalch equation and considering 95% of confidence using Prism 4 for Windows, GraphPad Software, San Diego, CA.
Fig. 9.
pKa value of Cp residue of MfOhrdel. Monobromobimane alkylation was performed using 4 µM pre-reduced MfOhrdel and 8 µM monobromobimane in the presence of 15 mM acetic acid, 15 mM MES [2-(N-Morpholino Ethanesulfonic Acid] and 30 mM Tris-HCl buffer adjusted to pH from 3.5 until 9.0. The determined pKa value of Cp from MfOhrdel was 5.7 (±0.1), according to non-linear regression using Henderson-Hasselbalch equation (GraphPad Software). This figure is representative of two independent sets of experiments.
2.12. Affinity purification of X. fastidiosa anti-Ohr serum
Since Ohr proteins present high structural similarity among themselves, we purified serum antibodies raised against Ohr from X. fastidiosa [4] to increase their specificity towards eukaryotic Ohr. Firstly, the His-tag from recombinant MfOhr was digested with thrombin following manufacturer instructions (Thrombin CleanCleave™ Kit, SIGMA Aldrich). The cleaved protein was covalently attached to a CNBr sepharose resin and stored in PBS pH 8.0. Then, the MfOhr sepharose beads were incubated with 2 mL of serum anti-XfOhr diluted in 8 mL of PBS pH 8.0 during 2 h at 4 °C. After, the beads were extensively washed with PBS pH 8.0 and antibodies were eluted from the resin with 2 mL of 0.2 M glycine pH 2.8. The samples eluted (500 µL fractions) were immediately neutralized with 20 μL of 3 M Tris-HCl pH 8.8 and 100 µL of 3 M KCl.
2.13. Protoplastization of M. fijiensis mycelia
Approximately 106 conidia x mL−1 of M. fijiensis Mf_1 were inoculated in 100 mL of PDB medium supplemented with 34 µg/mL of chloramphenicol and incubated during 36 h at 30 °C under agitation (200 rpm). Cell walls of mycelia were digested with a mix of enzymes Lallzyme MMX (15g/L), lysing enzymes from Trichoderma harzianum (5g/L) (Sigma-Aldrich cat # L1412) and BSA (10g/L) in the presence of 50 mL of solution 1 (0.8 M ammonium sulfate, 0.1 M citric acid, pH 6), 50 mL of solution 2 (1% (w/v) yeast extract, 2% (w/v) sucrose, pH 6) and 26 mL of 1 M MgSO4 and incubated during 5 h at 30 °C under gently agitation (100 rpm). Digested mycelia were filtered through a glass wool in order to separate protoplasts from cell debris and centrifuged 4000 rpm during 10 min at 4 °C.
2.14. Subcellular fractionation
The subcellular fractionation was adapted from [30]. Briefly, To isolate mitochondria, protoplasts were suspended in 10 mL of SHE buffer (0.6 M sorbitol, 20 mM HEPES and 1 mM EDTA) supplemented with 1 mM PMSF and mechanical disrupted after 50 strokes on ice using a dounce homogenizer. The lysed protoplasts solution, that represented the total fraction (TF), was centrifuged at 3800 rpm for 7 min at 4 °C and the supernatant was reserved. The same procedure was repeated three times to wash away all cellular debris and some organelles like nuclei and the supernatant collected was submitted to a final centrifugation step at 16,000xg during 10 min at 4 °C to separate supernatant (cytoplasmic fraction, CF) from pellet (enriched mitochondrial fraction). The enriched mitochondrial fraction (EMF) was suspended in 200 µL of SHE buffer and all collected fractions were stored at −80 °C.
2.15. MfOhr subcellular localization
To determine the subcellular localization of MfOhr, 15 or 30 µg of protein from extracts that correspond to the total fraction (TF), cytoplasmic fraction (CF) and enriched mitochondrial fraction (EMF) were separated by SDS-PAGE and the proteins were transferred to a nitrocellulose membrane. The membrane was stained with Ponceau S to check the amount of loaded proteins and incubated with antibodies that specifically target proteins from the cytoplasm (anti-PGK-1, phosphoglycerate kinase 1, Nordic BioSite cat. number BT-BS6691) or mitochondria (anti-COX IV, cytochrome c oxidase subunit IV, Abcam ab14744).
3. Results
3.1. Ohr/OsmC homologs data mining and their taxonomic distribution among eukaryotes
Previously, Ohr and OsmC enzymes were thought to be present only in bacteria [2], [6]. Our searches for homologs using delta-BLAST [15] and jackhmmer [16] in NCBI nr database revealed the existence of 392 eukaryotic proteins belonging to the Ohr/OsmC family (Table S1). Successive alignments of all of these eukaryotic sequences allowed us to ascertain that 189 of these sequences belong to Ohr subfamily and 5 sequences belong to the OsmC subfamily. We also identified a third group of Ohr/OsmC sequences, named here as Ohr-like, that comprise 198 proteins from eukaryotic organisms (Table S1) and that await further characterization.
Ohr, OsmC and Ohr-like proteins share a conserved pair of catalytic cysteines separated by approximately 60 amino acid residues in the primary sequence (Fig. 1) that, therefore, represents a hallmark feature of Ohr/OsmC of family proteins. Two additional residues (an Arg and a Glu) required for the peroxidatic activity [2], [4], [6], [14] are both fully conserved in Ohr and OsmC subfamilies (Fig. 1A and B) but are absent in Ohr-like proteins (Fig. 1C). This conserved Glu residue is located at the same position in the primary sequences of Ohr and OsmC proteins, while the conserved Arg residue is located in the first loop between the 1st and 2nd β-sheets for Ohr proteins (Fig. 1A); and in the third loop between the 3rd β-sheet and the 1st α-helix for OsmC (Fig. 1B). Although the conserved Arg residue is present at different positions in the primary sequences of Ohr and OsmC enzymes, in the tertiary structures they occupy a similar orientation between the conserved Glu and Cp [1], [12].
Fig. 1.
Multiple sequence alignment of selected members from Ohr, OsmC and Ohr-like subfamilies. The sequences were aligned using L-INS-I algorithm of MAFFT [13]. For each subfamily, sequences from different bacteria phyla were aligned with selected Ohr eukaryotic sequences. (A) For Ohr, 4NOZ secondary structure from Burkholderia cenocepacia J2315 (Bc_Ohr) was used to guide the alignment. Green and red arrows denote catalytic Arg and Glu. The selected Ohr sequences of prokaryotes were: Rs_Ohr from Ralstonia solanacearum UW551, Sa_Ohr from Stigmatella aurantiaca, Pg_Ohr from Polymorphum gilvum, Pa_Ohr from Propionibacterium acnes, Cc_Ohr from Clostridium carboxidivorans, Sp_Ohr from Sphingobacterium paucimobilis, Ds_Ohr from Deinococcus swuensis, Mr_Ohr from Mastigocladopsis repens, Kr_Ohr from Ktedonobacter racemifer and Ma_Ohr from Mycoplasma alligatoris. The selected Ohr sequences of eukaryotes were: Pp_Ohr from Physcomitrella patens, Kf_Ohr from Klebsormidium flaccidum, Mf_Ohr from Mycosphaerella fijiensis CIRAD86, Pm_Ohr from Pseudocercospora musae, Fo_Ohr from Fusarium oxysporum f. sp. cubense race 4, Ai_Ohr from Aphanomyces invadans, Bd_Ohr from Batrachochytrium dendrobatidis JEL423, Cc_Ohr from Calocera cornea HHB12733 Cc_1, Rt_Ohr from Rhodotorula toruloides ATCC 20409, Me_Ohr from Mortierella elongata AG-77, Mv_Ohr from Mortierella verticillata NRRL 6337. (B) For OsmC, 1QLM secondary structure from Escherichia coli (Ec_OsmC) was used to guide the alignment. Green and red arrows denote catalytic Arg and Glu. The selected OsmC sequences of prokaryotes were: Vp_OsmC from Variovorax paradoxus, Ab_OsmC from Azospirillum brasilense, Bb_OsmC from Bdellovibrio bacteriovorus, Sa_OsmC from Streptomyces avermitilis, Fa_OsmC from Flavobacterium aquatile, Kr_OsmC from Ktedonobacter racemifer, Df_OsmC from Deinococcus frigens, Ll_OsmC from Lactococcus lactis, Fm_OsmC from Fischerella muscicola and Vs_OsmC from Verrucomicrobium spinosum. The selected OsmC sequences of eukaryotes were: Pp_OsmC from Polysphondylium pallidum PN500, Dp_OsmC from Dictyostelium purpureum, As_OsmC from Acytostelium subglobosum LB1 and Dd1_OsmC from Dictyostelium discoideum. (C) Selected Ohr-like sequences deposited in PDB database were aligned with selected Ohr-like members from eukaryotic counterparts. For Ohr-like, secondary structure 2PN2 from Psychrobacter arcticus 273-4 (Pa_Ohr_like) was used to guide the alignment. The selected OsmC sequences of prokaryotes were: Aa_Ohr_like from Aquifex aeolicus, Js_Ohr_like from Jannaschia sp., Ll_Ohr_like from Lactobacillus casei, Tm_Ohr_like from Thermotoga maritima, Ta_Ohr_like from Thermoplasma acidophilum. The selected Ohr-like sequences of eukaryotes were: Mp_Ohr_like from Micromonas pusilla CCMP1545, Ep_Ohr_like from Exaiptasia pallida, Tv_Ohr_like from Trichomonas vaginalis G3, To_Ohr_like from Thalassiosira oceanica, Ai_Ohr_like from Aphanomyces invadans, Mf_Ohr_like from Mychosphaerella fijiensis CIRAD86, Fo_Ohr_like from Fusarium oxysporum f. sp. cubense race 4, Cc_Ohr_like from Calocera cornea HHB12733, Rt_Ohr_like from Rhodotorula toruloides ATCC 204091 and Co_Ohr_like from Capsaspora owczarzaki ATCC 30864. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Members of Ohr/OsmC family were detected in all eukaryotic groups, except Metazoa (Fig. 2A), considering the Tree of Life and taxonomy proposed by [31]. The largest number of Ohr/OsmC homologs was observed in Fungi (76% or 300/392 of sequences), mainly in the Ascomycota and Basidiomycotate phyla. Other microbial eukaryotes from a wide range of clades contain about 16% (63/392 of sequences) of Ohr/OsmC homologs, such as Euglenozoa; Amebozoa; Metanomada (Trichomonas vaginalis); Heterolobosea (Naegleria gruberi strain NEG-M); and non-metazoan Holozoa, such as Choanoflagellida (Salpingoeca, Monosiga), Ichthyosporea (Sphaeroforma) and Filasterea (Capsaspora). Close to 5% (20/392) of all Ohr/OsmC sequences were found in the SAR (Stramenopiles, Alveolata and Rhizaria) clade, such as Alveolata (Tetrahymena, Ichthyophthirius and Vitrella brassicaformis CCMP3155), Stramenopiles (Aphanomyces, Saprolegnia, Thalassiosira and Nannochloropsis) and Rhizaria (Reticulomyxa filosa) groups. Among the photosynthetic eukaryotes, Cryptista (Guillardia theta CCMP2712), Haptophyceae (Emiliania huxleyi CCMP1516), and non-vascular plants encode homologs of Ohr/OsmC genes in their genomes. Genes from non-vascular plants correspond to almost 2% of Ohr/OsmC homologs and were detected in the Chlorophyta (Chlorella and Micromonas), Bryophyta (Physcomitrella patens), Marchantiophyta (Marchantia polymorpha) and Streptophyta (Klebsormidium flaccidum).
Fig. 2.
RAxML maximum likelihood phylogenetic tree for eukaryotic members of the Ohr/OsmC family. (A) Unrooted phylogenetic tree of 392 eukaryotic sequences plus 78 selected Ohr sequences from bacteria of distinct phyla. (B) Unrooted phylogenetic tree of 186 Ohr eukaryotic sequences plus 78 selected Ohr sequences from bacteria of distinct phyla. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
We also retrieved sequences from Metazoa, including sequences from nematode, Trichuris trichiura (GenBank accession CDW57322.1 and described in [22] as a member of Ohr/OsmC family), insect, Drosophila eugracilis (XP_017066882), crustacean, Daphnia magna (KZS01297) and sea anemona Exaiptasia pallida (KXJ04390). However, these sequences have extremely high amino acid identity to sequences from Enterococcus (100%), Acetobacter (100%), Burkholderia (98%) and Oceanospirillum (97%), respectively. Furthermore, there is no evidence of a signal peptide sequence in these animal proteins and it is most likely that these sequences are spurious, being derived from DNA from symbiotic bacteria or sample contamination [32], [33]. Therefore, we did not include these sequences in our analysis.
3.2. Distribution of Ohr, OsmC and Ohr-like subfamilies homologs among eukaryotes
To gain insights on the evolutionary relationships among Ohr, OsmC and Ohr-like sequences from different eukaryotic phyla, we analyzed the data retrieved using Maximum Likelihood (ML) phylogenetic inference. This resulted in a tree with well-defined clades bearing the signatures of each family (Fig. 2A and B). The occurrences of Ohr, OsmC and Ohr-like proteins among eukaryotic phylogenetic groups are quite distinct. While Ohr and OsmC homologs are more restricted to Fungi and cellular slime molds, respectively, Ohr-like homologs are widespread among various eukaryotic groups (Fig. 2A).
The 189 sequences from Ohr subfamily compose a well-defined group that is isolated from bacterial counterparts and are mostly present in Fungi of the Ascomycota (87/189) and Basidiomycota (98/189 sequences) phyla (Fig. 2B). In contrast, the five members of the OsmC subfamily are grouped into a single monophyletic clade restricted to the Dictyostelia order (cellular slime molds) (Fig. 2A).
Concerning the Ohr-like subfamily (198 sequences), most of them are present in Ascomycota (90 sequences), while only 16 sequences could be found in Basidiomycota. Ohr-like homologs are also abundant among Euglenozoa (48 sequences), but can also be found in non-metazoan Holozoa, such as Sphaeroforma arctica JP610 (two Ohr-like paralogues) and Capsaspora owczarzaki (one Ohr-like), some of the closest unicellular relatives of multicellular animals [34]. Interestingly, a separated analysis that included only Ohr homologs and some selected bacterial Ohrs revealed that five eukaryotic Ohr homologs grouped within the bacterial Ohr group (Fig. 2B). These sequences were encoded by genes from different species of Basidiomycota, such as Calocera (C. cornea and C. viscosa) and Dacryopinax (represented by purple circles in the Fig. 2B) and Stramenopiles Aphanomyces invadans and A. astaci (represented by light green triangles in Fig. 2B). These latter sequences are likely examples of very recent horizontal gene transfer events from bacterial lineages, given the absence of introns in organisms such as Aphanomyces and their low levels of similarity of sequences from organelles and organelle related bacterial lineages.
3.3. Genomic configurations of ohr/osmC genes in eukaryotic organisms
Several eukaryotic genomes present more than one member of the Ohr/OsmC family, being arranged in highly variable configurations (Fig. 3). For instance, some fungi microorganisms present two or three ohr paralogues in their genomes (Fig. 3A and B), some located near to each other, in some cases the two gene are even neighbors (Fig. 3A), suggesting the occurrence of gene duplication events. Besides fungi, microorganisms that contain more than one gene of the Ohr/OsmC family are: M. fijiensis presenting one ohr and one ohr-like gene (Fig. 3C); Trichomonas vaginalis with four ohr-like genes (as also described by [22]) and D. discoideum AX4 containing one copy of osmC and one copy of an ohr-like gene (Fig. 3D). On the other hand, most of the other genomes encode only a single homolog of the ohr subfamily, as is the case of the moss P. patens (Fig. 3E).
Fig. 3.
Genomic arrangements of genes of the Ohr/OsmC family present in eukaryotes.
We also observed that many eukaryotic ohr/osmC genes present introns, however their evolutionary significance is still elusive. The complete list of ohr, osmC or ohr-like genes (and their predicted introns), as well as their abundance in each specie, is presented in table S 1.
3.4. Recombinant eukaryotic Ohr and OsmC have peroxidase activity
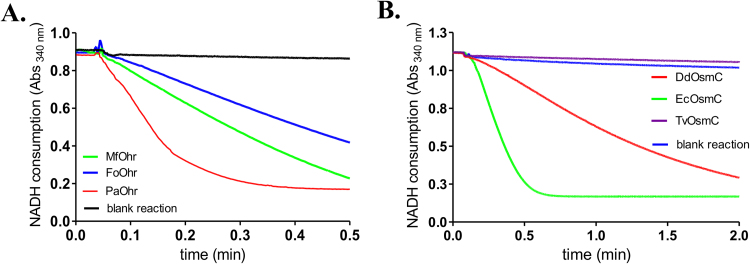
To verify whether Ohr and OsmC homologs present in eukaryotic organisms also display lipoyl peroxidase activity as their bacterial counterparts, we obtained recombinant Ohr proteins from M. fijiensis (MfOhr) and from F. oxysporium (FoOhr) and recombinant OsmC from Dictyostelium discoideum (DdOsmC), expressed in E. coli. All the selected fungal Ohr enzymes reduced tBOOH (Fig. 4A). The FoOhr reduced tBOOH at lower rates when compared with MfOhr and both proteins were less efficient peroxidases than their bacterial counterpart (PaOhr). Considering eukaryotic OsmC, DdOsmC presented about half of NADH consumption compared to the bacterial OsmC counterpart (E. coli BW25113) in the experimental conditions analyzed (Fig. 4B).
We chose MfOhr for further characterization as it displayed the highest activity among the eukaryotic proteins studied.
3.5. MfOhr enzymatic properties are similar to the bacterial Ohrs
Peroxidase activity of MfOhr was specifically supported by dithiols (DTT and DHLA) and not by monothiols (GSH and β-mercaptoethanol) (Fig. 5) and thus similar to bacterial Ohr enzymes [3].
Fig. 5.
Thiol specificity of MfOhr peroxidase activity. The amount of tBOOH remaining in solution after reaction was determined by the FOX assay. Reactions were initiated by addition of thiol compounds and terminated by addition of 20 µL of HCl (5 M) into 100 µL reaction mixtures. Reactions were carried out in 50 mM Tris-HCl buffer pH 7.4 in the presence of MfOhr (0.5 μM), sodium azide (1 mM) and DTPA (0.1 mM). The tested reducing agents were: 5 mM reduced glutathione (GSH), 10 mM 2-mercaptoethanol, 0.5 mM Dithiothreitol (DTT) or Dihydrolipoamide (DHLA).
We also analyzed MfOhr's specificity towards the oxidizing substrate, since it is well established that bacterial Ohr enzymes present higher preference for organic hydroperoxides over H2O2 [2], [3], [4], [10]. Indeed, the specific activity of MfOhr for CuOOH was almost six times higher than the values presented for tBOOH and almost 500 times higher in comparison to H2O2 (Fig. 6). In conclusion, similarly to bacterial Ohr, MfOhr also displayed greater affinity for more hydrophobic substrates.
Fig. 6.
Specific activities of MfOhr and MfOhrdel towards CuOOH, tBOOH and H2O2. The values were calculated for the wild type (173 amino acids, MfOhr) or truncated (140 amino acids, MfOhrdel) versions of Mf_1 Ohr, using the lipoamide/ lipoamide dehydrogenase coupled assay. Initial rates were obtained from the linear portion of the curves from reactions performed at 0.075, 0.1, 0.25 and 0.5 µM of enzyme for tBOOH, 0.025, 0.05, 0.075 and 0.15 µM of enzyme for CuOOH and 10, 12 and 15 µM of enzyme for H2O2.
We also evaluated the specific activities of a processed version of Ohr (MfOhrdel), in which the first 33 amino acids residues were removed, as it is well established that the N-terminal sequence is proteolytically cleaved during import of the proteins into mitochondria. Both versions of the MfOhr (processed versus not processed) display similar specific activities values for each hydroperoxide tested (Fig. 6), suggesting that the N-terminal extension does not influence protein activity. In the following assays, we then chose to use the processed version of MfOhr (MfOhrdel).
Since tBOOH and CuOOH are synthetic compounds, we decided to evaluate the ability of MfOhr to reduce more complex hydroperoxides, such as fatty acid hydroperoxides. In fact, we recently described that hydroperoxides derived from oleic, linoleic and arachidonic fatty acids are the biological substrates of bacterial Ohr, at least for enzymes from P. aeruginosa and X. fastidiosa [11]. Therefore, we determined the second order rate constant for the reaction between the reduced MfOhrdel and linoleic acid hydroperoxide (LAOOH) by employing a competitive approach that follows redox dependent changes in the AhpE intrinsic fluorescence [11]. Like the bacterial enzymes, MfOhrdel displayed an extraordinarily high rate constant (3.2 (±2.1)×108 M−1 s−1) for LAOOH reduction (Fig. 7A and B).
Fig. 7.
Kinetics of LAOOH reduction by MfOhr. A. Reaction of MfOhrdel with LAOOH was investigated by a competitive assay following the intrinsic tryptophan fluorescence emission (λex=290 nm and λem=340 nm) of 2 µM AhpE [11]. The red and yellow lines represent the emission fluorescence of AhpE in its reduced (no LAOOH) and oxidized (added 1.8 µM LAOOH) states; in the absence of MfOhrdel. In the competition assay, reduced AhpE was mixed with 0.5 (green line), 1 (blue line), 1.5 (brown line) and 2 µM (black line) of reduced MfOhrdel in the presence of 1.8 µM LAOOH. B. The fraction of oxidized AhpE (AhpEOx) decreases with increasing amount of MfOhrdel. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
3.6. Single Cys mutants of MfOhr do not have detectable peroxidase activity
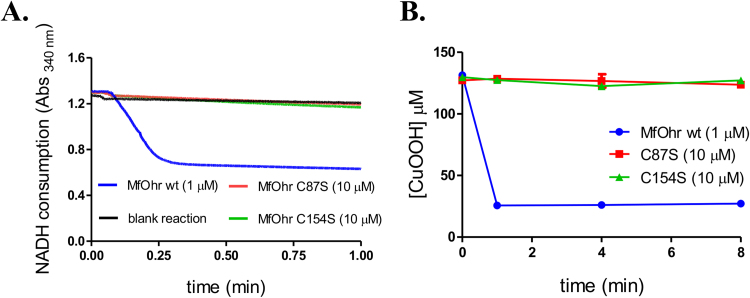
To evaluate the catalytic role of Cys residues of MfOhr, we generated single mutants for each residue. Both mutants lost their peroxidase activity as assessed by lipoamide/lipoamide dehydrogenase coupled assay and FOX assay (Fig. 8). Similar results were observed for Ohr from X. fastidiosa [3]. These results suggest the Cr might have a role in activating Cp for hydroperoxide reduction.
Fig. 8.
Comparison of the peroxidase activities of MfOhrdel and the C87S and C154S mutants. A. Lipoamide/lipoamide dehydrogenase coupled assay. The reactions were performed at 37 °C with 1 µM MfOhrdel (blue line) or 10 µM mutant proteins (C87S, red line or C154S, green line), in the presence of 50 µM reduced lipoamide, 100 µM DTPA, 0.5 µM XfLpd and 200 µM NADH in 50 µM sodium phosphate pH 7.4. Reactions were initiated by addition of 200 µM CuOOH. Blank reaction (black line) was performed without enzyme. B. The consumption of CuOOH was monitored during 8 min using FOX assay. The reactions were carried out in the presence of 1 µM (MfOhrdel) or 10 µM (C154S or C86S) enzymes. The control reactions for each tested hydroperoxide (enzyme+peroxide without DTT) and (hydroperoxide+DTT without enzyme) are not showed here. The figure is representative of at least two independent sets of experiments. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
3.7. Determination of cysteine's pKa values
All thiol peroxidases so far described carry a reactive Cys, the so called Cp, whose thiolate group displays an acidic pKa [35]. Therefore, we decided to determine the pKa value of CP of MfOhrdel by the monobromobimane alkylation method [29]. The curve that best fitted to the experimental data was obtained by nonlinear regression (Handerson-Hasselbach equation) and resulted in a pKa value of 5.7±0.1 for the Cp residue (Fig. 9). This value is very similar to that previously described for bacterial type Ohr, 5.3±0.1 [35]. We were unable to detect the pKa of resolving Cys probably because this residue is deeply buried in the polypeptide backbone, as observed for bacterial Ohr enzymes, whose structures were elucidated [25]. Indeed, MfOhr C87S did not display any fluorescence upon mBrB treatment, supporting the hypothesis that Cr (Cys 154) is inaccessible to this alkylating agent under the experimental conditions employed here (data not shown).
3.8. MfOhr intramolecular disulfide bond formation upon hydroperoxide treatment
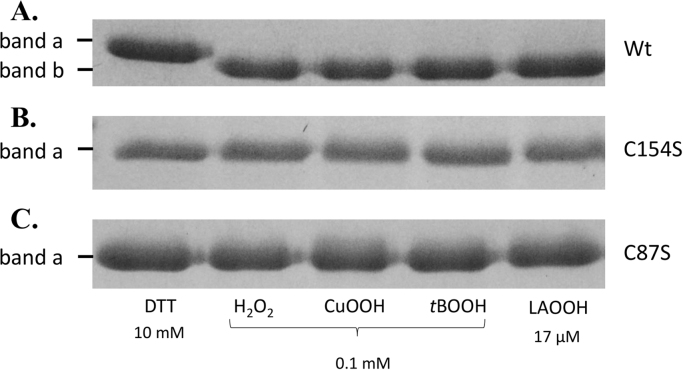
We next studied the thiol redox state of Cys residue in response to hydroperoxides by non-reducing SDS-PAGE, since the intramolecular disulfide bond of Ohr enzymes can be detected due to its lower hydrodynamic volume as a band (band b) that migrates faster than the reduced state (band a) [14]. Wt, C87S and C154S MfOhr were exposed to reducing (10 mM of DTT) or oxidative conditions (0.1 mM of CuOOH, tBOOH or H2O2 and 0.017 mM of LAOOH) during 1 h at 37 °C. For the Wt MfOhr, we observed the appearance of band b upon oxidation as expected since it corresponds to the intramolecular disulfide (Fig. 10A). Band b was not observed when Cp or Cr residues were independently substituted by serine residues. In this case, a single band (band a) was observed that migrated equally regardless of conditions (Fig. 10B and C).
Fig. 10.
Non-reducing SDS-PAGE gels showing the effect of DTT and hydroperoxide treatments on MfOhrdel (A), MfOhrdel C154S (B) and MfOhrdel C87S (C). 10 µM of each protein were incubated during 1 h at 37 °C with 10 mM DTT, 0.1 mM H2O2, CuOOH or tBOOH or 17 µM linoleic acid hydroperoxide (LAOOH). All reactions were carried out in a buffer containing 0.5 M NaCl, 20 mM sodium phosphate pH 7.4 and 1 mM DTPA. Immediately after DTT or hydroperoxides treatments, all the samples were alkylated with NEM (100 mM) for 1 h at room temperature to avoid oxidation artefacts due to protein denaturation by SDS.
Therefore, again as bacterial enzymes, MfOhr is oxidized to a stable intramolecular disulfide upon oxidation by hydroperoxides.
3.9. Is MfOhr targeted to mitochondria?
Careful analysis of eukaryotic Ohr/OsmC sequences revealed that in most cases these sequences are longer than bacterial Ohr sequences (Table S1). The eukaryotic sequences contain an N-terminal extension that could harbor a signal peptide sequence for organellar localization or for extracellular secretion. Indeed, analysis of all 392 sequences by the TargetP and [28] and Mitofates [36] methods predicted that most of these proteins are addressed to mitochondria or another organelle (p>90%) (Table S1).
To experimentally verify if MfOhr is in fact a mitochondrial protein, we performed the subcellular fractionation of M. fijiensis protoplast cells, followed by western blot analysis. Initially, the affinity of purified Ohr antibody raised against bacterial Ohr was shown to be able to detect recombinant MfOhrdel (Fig. S2). However, MfOhr could not be detected in whole extracts of M. fijiensis mycelia grown in PDB medium, even when high amounts of total protein (200–300 µg) were employed. In contrast, a strong signal was observed in the enriched mitochondrial fraction (Fig. 11B). This is likely due to a dilution effect as the mitochondria occupy only a small fraction of the whole cellular mass. Indeed, COX IV, a well-established and abundant mitochondrial protein, was detected in whole extract at significantly lower levels than in the enriched mitochondrial fraction (Fig. 11D). As another control, the cytoplasmic protein PGK-1 was present in the total and cytoplasmic fraction but not in the mitochondrial fraction (Fig. 11C). Taken together, these results confirm the in silico prediction that MfOhr is a mitochondrial protein.
Fig. 11.
Western blots of total (TF), cytosolic (CF) and enriched mitochondria (EMF) fractions of protoplasts cells of M. fijiensis Mf_1. Fractions of M. fijiensis protoplasts are described in Material and Methods. A. Loading control (ponceau staining) of cellular fractions. After western blot, membrane was probed with affinity purified MfOhr polyclonal antibody (B); PGK-1, a cytoplasmic marker (C) and COX-IV, a mitochondrial marker (D), respectively.
4. Discussion
Initially thought to be exclusively found in prokaryotes, Ohr proteins have been described as the main enzymatic system involved in bacterial defense against organic hydroperoxides [5], [7], [9], [10], [35], [37]. We show here that members of the Ohr/OsmC family are also present in several eukaryotic clades and are especially common among species of Fungi. To validate these in silico observations, biochemical characterization of selected proteins was carried out and our results indicated that these eukaryotic peroxidases are enzymatically similar to their bacterial counterparts.
Furthermore, we also demonstrated, not only for the Ohr subfamily but also for almost all eukaryotic Ohr/OsmC proteins, that the N-terminal signal sequence is predicted to localize to mitochondria or another organelle, such as the peroxisome or chloroplasts (Table S1). The presence of Ohr in mitochondria is in agreement with the fact that the proposed reducing system, i. e. lipoylated proteins from α-ketoacid dehydrogenase complexes [4], is also present in mitochondria. These findings are also consistent with the observation that mitochondria is a major source of endogenous oxidants [38], [39]. It is reasonable to think that for eukaryotes, the peroxidase activity of Ohr may be related with detoxification of endogenous sources of hydroperoxides and might be not involved in defense towards exogenous insults of ROS, as it is currently proposed for bacteria [11], [40], [41].
It is noteworthy, that several species presenting Ohr homologs in eukaryotes are non-vascular plant or animal pathogens (Table S1), such as M. fijiensis, the causative agent of Black Sigatoka, the most important disease of banana and plants worldwide [42]. Remarkably, when we looked at the sequenced genomes currently available, we found that Ohr enzymes are completely absent in animals and vascular plants, although these organisms are hosts for microbial pathogens, thus making Ohr proteins a convenient target for drug development.
OsmC homologs also display thiol dependent peroxidase activity and show preference for organic hydroperoxides [43], [44], [45], [46]. We also found OsmC enzymes in eukaryotes, but in this case the taxonomic distribution is restricted to some species within Dictyostelia. Interestingly, the osmC gene from D. discoideum AX4 was previously suggested to have been acquired via lateral gene transfer [47] but never had its peroxidase activity reported. To our knowledge, this is the first study that actually determined the peroxidase activity for an OsmC enzyme from Dictyostelium or any other eukaryotic organism.
Classification of proteins in the Ohr/OsmC family that do not belong to the Ohr and OsmC subfamilies were already described in the literature [1]. However, in these studies low numbers of sequences were analyzed. Here, Ohr-like sequences (not belonging to Ohr or OsmC subfamilies) were grouped together (Fig. 2), but we are aware that further studies are required for proper classification of these enzymes and an analysis including all available sequences of Ohr-like enzymes is currently under investigation in our laboratory.
Recently, the peroxidase activity of a Ohr-like enzyme from T. vaginalis was described [22] (Genbank accession no. XP_001323255). This protein was named TvOsmC, although it does not have the conserved Arg and Glu residues characteristic of the OsmC subfamily (Fig. 1B and C). In our experiments, TvOsmC did not display peroxidase activity (Fig. 4B and S1, respectively). In contrast, the peroxidase activity of TvOsmC was detected by Nývltová et al., [19]. Possibly, L and H (lipoylated) proteins from hydrogenossomes of T. vaginalis are required to support the peroxidase activity of this enzyme.
In summary, we showed that Ohr/OsmC proteins, mostly present in bacteria, also occur among eukaryotes, and are mostly targeted to organellar compartments. Although it is currently proposed that these genes were acquired from prokaryotes through lateral gene transfer events [22], [47] and we did describe some cases for recent transfer events, our phylogeny cannot confirm nor negate the hypothesis of ancient transfers due to low bootstrap values in the deep branches of the Ohr/OsmC tree (Fig. 2A). On the other hand, to assume that members of the eukaryotic Ohr subgroup, which only includes genes from fungi, originate from endosymbiont-derived genes present in the last common ancestor of all eukaryotes would require multiple gene losses at the root of different eukaryotic lineages [48], [49]. A more parsimonious explanation is that lateral gene transfer from bacteria to a lineage close to the common ancestor of extant fungal lineages was responsible for the unique presence of Ohr among fungi. As sequence databases grow and more sequences are added, further evolutionary studies will undoubtedly help improve our understanding of the origin and evolution of proteins from the Ohr/OsmC family across the many branches in the tree of life.
Acknowledgements
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo – FAPESP, process 2013/07937-8 (Redox Processes in Biomedicine -REDOXOMA). D.A.M was recipient of a fellowship from FAPESP, process 2012/21722-1. We thank Dr. Miguel J. Beltrán-García from Departamento de Química, Universidad Autónoma de Guadalajara, Guadalajara, Jalisco México and Dr. Aline Maria da Silva and Dr. Layla Farage Martins from Departamento de Bioquímica, Instituto de Química, Universidade de São Paulo, São Paulo, Brazil for kindly provide the M. fijiensis Mf_1 and D. discoideum AX4 strain, respectively.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2017.03.026.
Contributor Information
D.A. Meireles, Email: meireles@ib.usp.br.
L.E.S. Netto, Email: nettoles@ib.usp.br.
Appendix A. Supplementary material
Fig. S1.
Peroxidase activity of TvOsmC and DdOsmC assessed by oxidized DTT assay. Reactions were carried out in the presence of 5 µM DdOsmC (blue line) or TvOsmC (green line), sodium phosphate pH 7.4 (100 mM), DTPA (1 mM) and CuOOH (2 mM), and started by adding of 10 mM reduced DTT, approximately 20 s after the mixing of the other reactants. As positive control of reaction, the same assay was made using 5 µM EcOsmC (red line). The blank reaction (black line) was performed without any enzyme addition. The figure is representative of at least two independent sets of experiments.
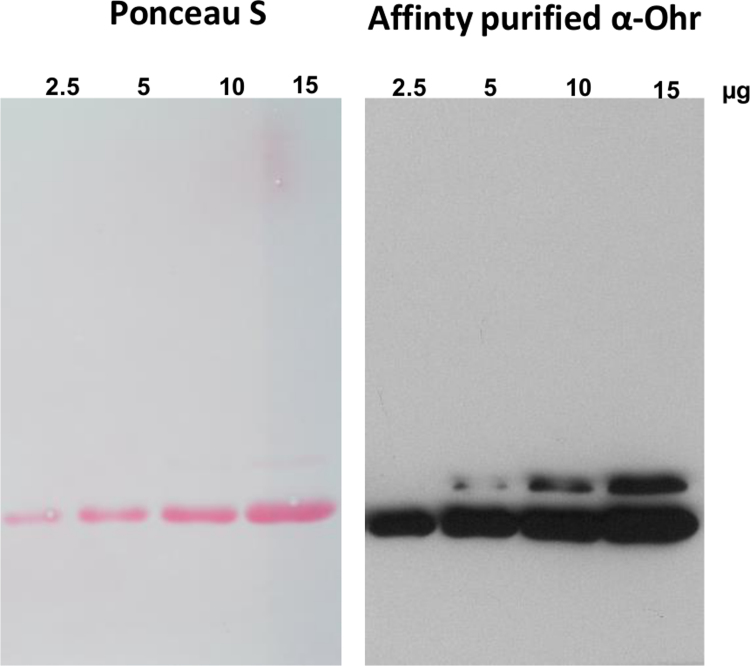
Fig. S2.
Affinity purified XfOhr serum recognizes MfOhrdel. The gel was loaded with increasing concentrations of His-cleaved MfOhrdel. Details of cleavAge procedure are shown in M&M.
Fig. S3.
Whole membrane from experiment depicted in the Fig. 10.
Supplementary material Fig S4 Full rectangular view of phylogenetic tree showed in Fig. 1. Bootstraps support values above 50 are showed on the nodes.
Supplementary material Table S1 – List of Genbank Accession Numbers (NCBI) of retrieved eukaryotic sequences that belong to the Ohr/OsmC family. For each sequence, some features such as length, taxonomy and the signal peptide prediction are also listed.
References
- 1.Shin D.H., Choi I.-G., Busso D., Jancarik J., Yokota H., Kim R., Kim S.-H. Structure of OsmC from Escherichia coli: a salt-shock-induced protein. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:903–911. doi: 10.1107/S0907444904005013. [DOI] [PubMed] [Google Scholar]
- 2.Lesniak J., Barton W.A., Nikolov D.B. Structural and functional characterization of the Pseudomonas hydroperoxide resistance protein Ohr. EMBO J. 2002;21:6649–6659. doi: 10.1093/emboj/cdf670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cussiol J.R.R., Alves S.V., de Oliveira M.A., Netto L.E.S. Organic hydroperoxide resistance gene encodes a thiol-dependent peroxidase. J. Biol. Chem. 2003;278:11570–11578. doi: 10.1074/jbc.M300252200. [DOI] [PubMed] [Google Scholar]
- 4.Cussiol J.R.R., Alegria T.G.P., Szweda L.I., Netto L.E.S. Ohr (organic hydroperoxide resistance protein) possesses a previously undescribed activity, lipoyl-dependent peroxidase. J. Biol. Chem. 2010;285:21943–21950. doi: 10.1074/jbc.M110.117283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mongkolsuk S., Praituan W., Loprasert S., Fuangthong M., Chamnongpol S. Identification and characterization of a new organic hydroperoxide resistance ( ohr) gene with a novel pattern of oxidative stress regulation from Xanthomonas campestrispv. Phaseoli Identification and characterization of a new organic hydroperoxide resi. J. Bacteriol. 1998;180:2636–2643. doi: 10.1128/jb.180.10.2636-2643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atichartpongkul S., Loprasert S., Vattanaviboon P., Whangsuk W., Helmann J.D., Mongkolsuk S. Bacterial Ohr and OsmC paralogues define two protein families with distinct functions and patterns of expression. Microbiology. 2001;147:1775–1782. doi: 10.1099/00221287-147-7-1775. 〈http://www.ncbi.nlm.nih.gov/pubmed/11429455〉 [DOI] [PubMed] [Google Scholar]
- 7.T. Chuchue, W. Tanboon, J.M. Dubbs, P. Vattanaviboon, S. Mongkolsuk, B. Prapagdee, ohrR and ohr are the Primary Sensor/Regulator and Protective Genes Against Organic Hydroperoxide Stress in Agrobacterium Tumefaciens ohrR and ohr are the Primary Sensor/Regulator and Protective Genes against Organic Hydroperoxide Stress in Agrobacteria, 2006. doi: 〈http://doi.org/10.1128/JB.188.3.842〉. [DOI] [PMC free article] [PubMed]
- 8.Conter A., Gangneux C., Suzanne M., Gutierrez C. Survival of Escherichia coli during long-term starvation: effects of aeration, NaCl, and the rpoS and osmC gene products. Res. Microbiol. 2001;152:17–26. doi: 10.1016/s0923-2508(00)01164-5. 〈http://www.ncbi.nlm.nih.gov/pubmed/11281321〉 [DOI] [PubMed] [Google Scholar]
- 9.da Silva Neto J.F., Negretto C.C., Netto L.E.S. Analysis of the organic hydroperoxide response of chromobacterium violaceum reveals that OhrR is a cys-based redox sensor regulated by thioredoxin. PLoS One. 2012;7:e47090. doi: 10.1371/journal.pone.0047090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Si M., Wang J., Xiao X., Guan J., Zhang Y., Ding W., Chaudhry M.T., Wang Y., Shen X. Ohr protects corynebacterium glutamicum against organic hydroperoxide induced oxidative stress. PLoS One. 2015;10:e0131634. doi: 10.1371/journal.pone.0131634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alegria T.G.P., Meireles D.A., Cussiol J.R.R., Hugo M., Trujillo M., de Oliveira M.A., Miyamoto S., Queiroz R.F., Valadares N.F., Garratt R.C., Radi R., Di Mascio P., Augusto O., Netto L.E.S. Ohr plays a central role in bacterial responses against fatty acid hydroperoxides and peroxynitrite. Proc. Natl. Acad. Sci. USA. 2017;114:E132–E141. doi: 10.1073/pnas.1619659114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker L.M.S., Poole L.B. Catalytic mechanism of thiol peroxidase from Escherichia coli. J. Biol. Chem. 2003;278:9203–9211. doi: 10.1074/jbc.M209888200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.K.J. Nelson, S.T. Knutson, L. Soito, C. Klomsiri, B. Leslie, J.S. Fetrow, Analysis of the Peroxiredoxin Family: Using Active Site Structure and Sequence Information for Global Classification and Residue Analysis, vol. 79, 2012, pp. 947–964. doi: 〈http://doi.org/10.1002/prot.22936.analysis〉. [DOI] [PMC free article] [PubMed]
- 14.Oliveira M. a., Guimarães B.G., Cussiol J.R.R., Medrano F.J., Gozzo F.C., Netto L.E.S. Structural insights into enzyme-substrate interaction and characterization of enzymatic intermediates of organic hydroperoxide resistance protein from Xylella fastidiosa. J. Mol. Biol. 2006;359:433–445. doi: 10.1016/j.jmb.2006.03.054. [DOI] [PubMed] [Google Scholar]
- 15.Boratyn G.M., Schäffer A. a., Agarwala R., Altschul S.F., Lipman D.J., Madden T.L. Domain enhanced lookup time accelerated BLAST. Biol. Direct. 2012;7:12. doi: 10.1186/1745-6150-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finn R.D., Clements J., Eddy S.R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y., Niu B., Gao Y., Fu L., Li W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.A. Stamatakis, Stamatakis – 2014 – RAxML Version 8 a Tool for Phylogenetic Analysis and Post-analysis of Large Phylogenies, 2014, pp. 2010–2011. [DOI] [PMC free article] [PubMed]
- 20.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:msw054. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cornillon S., Foa C., Davoust J., Buonavista N., Gross J.D., Golstein P. Programmed cell death in Dictyostelium. J. Cell Sci. 1994;107:2691–2704. doi: 10.1242/jcs.107.10.2691. [DOI] [PubMed] [Google Scholar]
- 22.Nývltová E., Smutná T., Tachezy J., Hrdý I. OsmC and incomplete glycine decarboxylase complex mediate reductive detoxification of peroxides in hydrogenosomes of Trichomonas vaginalis. Mol. Biochem. Parasitol. 2015 doi: 10.1016/j.molbiopara.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 23.Gasteiger E B.A., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D. Protein Identification and Analysis Tools on the ExPASy Server. Proteom. Protoc. Handb. 2005:571–607. [Google Scholar]
- 24.Ellman G.L. Tissue sulfhydril groups. Arch. Biochem. Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 25.Jiang Z.Y., Hunt J.V., Wolff S.P. Ferrous ion oxidation in the presence of xylenol orange for detection of lipid hydroperoxide in low density lipoprotein. Anal. Biochem. 1992;202:384–389. doi: 10.1016/0003-2697(92)90122-n. 〈http://www.ncbi.nlm.nih.gov/pubmed/1519766〉 [DOI] [PubMed] [Google Scholar]
- 26.Discola K.F., de Oliveira M.A., Rosa Cussiol J.R., Monteiro G., B??rcena J.A., Porras P., Padilla C.A., Guimar??es B.G., Netto L.E.S. Structural aspects of the distinct biochemical properties of glutaredoxin 1 and glutaredoxin 2 from Saccharomyces cerevisiae. J. Mol. Biol. 2009;385:889–901. doi: 10.1016/j.jmb.2008.10.055. [DOI] [PubMed] [Google Scholar]
- 27.Bryk R. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-Like protein. Science. 2002;295(80-.):1073–1077. doi: 10.1126/science.1067798. [DOI] [PubMed] [Google Scholar]
- 28.Trujillo M., Ferrer-Sueta G., Radi R. Chapter 10 kinetic studies on peroxynitrite reduction by peroxiredoxins. Methods Enzymol. 2008;441:173–196. doi: 10.1016/S0076-6879(08)01210-X. [DOI] [PubMed] [Google Scholar]
- 29.Sardi F., Manta B., Portillo-Ledesma S., Knoops B., Comini M.A., Ferrer-Sueta G. Determination of acidity and nucleophilicity in thiols by reaction with monobromobimane and fluorescence detection. Anal. Biochem. 2013;435:74–82. doi: 10.1016/j.ab.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 30.Schagger H., Cramer W.A., Vonjagow G. Mitochondrial biogenesis and genetics Part A. Anal. Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- 31.Hinchliff C.E., Smith S.A., Allman J.F., Burleigh J.G., Chaudhary R., Coghill L.M., Crandall K.A., Deng J., Drew B.T., Gazis R., Gude K., Hibbett D.S., Katz L.A., Laughinghouse H.D., McTavish E.J., Midford P.E., Owen C.L., Ree R.H., Rees J.A., Soltis D.E., Williams T., Cranston K.A. Synthesis of phylogeny and taxonomy into a comprehensive tree of life. Proc. Natl. Acad. Sci. USA. 2015;112:201423041. doi: 10.1073/pnas.1423041112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koutsovoulos G., Kumar S., Laetsch D., Stevens L., Daub J., Conlon C. No evidence for extensive horizontal gene transfer in the genome of the tardigrade Hypsibius dujardini. Pnas. 2016;113:1–6. doi: 10.1073/pnas.1600338113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salter S.J., Cox M.J., Turek E.M., Calus S.T., Cookson W.O., Moffatt M.F., Turner P., Parkhill J., Loman N.J., Walker A.W. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schalchian-Tabrizi K., Minge M.A., Espelund M., Orr R., Ruden T., Jakobsen K.S., Cavalier-Smith T. Multigene phylogeny of Choanozoa and the origin of animals. PLoS One. 2008;3 doi: 10.1371/journal.pone.0002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de D., Meireles A., eronimo T.G., Alegria P., idigal Alves S.V., ani C.R., Arantes R., duardo L.E., Netto S. A 14.7 kDa protein from Francisella tularensis subsp. novicida (named FTN_1133), involved in the response to oxidative stress induced by organic peroxides, is not endowed with thiol-dependent peroxidase activity. PLoS One. 2014;9:e99492. doi: 10.1371/journal.pone.0099492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukasawa Y., Tsuji J., Fu S.-C., Tomii K., Horton P., Imai K. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell. Proteom. 2015;14:1113–1126. doi: 10.1074/mcp.M114.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fontenelle C., Blanco C., Arrieta M., Dufour V., Trautwetter A. Resistance to organic hydroperoxides requires ohr and ohrR genes in Sinorhizobium meliloti. BMC Microbiol. 2011;11:100. doi: 10.1186/1471-2180-11-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quinlan C.L., Goncalves R.L.S., Hey-Mogensen M., Yadava N., Bunik V.I., Brand M.D. The 2-oxoacid dehydrogenase complexes in mitochondria can produce superoxide/hydrogen peroxide at much higher rates than complex I. J. Biol. Chem. 2014;289:8312–8325. doi: 10.1074/jbc.M113.545301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J., Wang Zhi, Zhou Hui, Sheng Zhigang, Naseer Ying, Kan Nawar, Biao, Zhu Thiol-based switch mechanism of virulence regulator AphB modulates oxidative stress response in Vibrio cholerae. Clin. Exp. Immunol. 2016;102:939–949. doi: 10.1111/mmi.13524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reniere M.L., Whiteley A.T., Portnoy D.A. An in vivo selection identifies Listeria monocytogenes Genes required to sense the intracellular environment and activate virulence factor expression. PLoS Pathog. 2016;12:1–27. doi: 10.1371/journal.ppat.1005741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Churchill A.C.L. Mycosphaerella fijiensis, the black leaf streak pathogen of banana: progress towards understanding pathogen biology and detection, disease development, and the challenges of control. Mol. Plant Pathol. 2011;12:307–328. doi: 10.1111/j.1364-3703.2010.00672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saikolappan S., Das K., Sasindran S.J., Jagannath C., Dhandayuthapani S. OsmC proteins of Mycobacterium tuberculosis and Mycobacterium smegmatis protect against organic hydroperoxide stress. Tuberculosis. 2011;91(Suppl 1):S119–S127. doi: 10.1016/j.tube.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang W., Baseman J.B. Functional characterization of osmotically inducible protein C (MG_427) from Mycoplasma genitalium. J. Bacteriol. 2014;196:1012–1019. doi: 10.1128/JB.00954-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lesniak J., Barton W.A., Nikolov D.B. Structural and functional features of the Escherichia coli hydroperoxide resistance protein OsmC. Protein Sci. 2003;12:2838–2843. doi: 10.1110/ps.03375603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park S.-C., Pham B.P., Van Duyet L., Jia B., Lee S., Yu R., Han S.W., Yang J.-K., Hahm K.-S., Cheong G.-W. Structural and functional characterization of osmotically inducible protein C (OsmC) from Thermococcus kodakaraensis KOD1. Biochim. Biophys. Acta. 1784;(2008):783–788. doi: 10.1016/j.bbapap.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 47.Eichinger L., Pachebat J.A., Glöckner G., Rajandream M., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q., Tunggal B., Kummerfeld S., Madera M., Konfortov B.A., Rivero F., Bankier A.T., Lehmann R., Hamlin N., Davies R., Gaudet P., Fey P., Pilcher K., Chen G., Saunders D., Sodergren E., Davis P., Kerhornou A., Nie X., Hall N., Anjard C., Hemphill L., Bason N., Farbrother P., Desany B., Just E., Morio T., Rost R., Churcher C., Cooper J., Haydock S., van Driessche N., Cronin A., Goodhead I., Muzny D., Mourier T., Pain A., Lu M., Harper D., Lindsay R., Hauser H., James K., Quiles M., Babu M. Madan, Saito T., Buchrieser C., Wardroper A., Felder M., Thangavelu M., Johnson D., Knights A., Loulseged H., Mungall K., Oliver K., Price C., Quail M.A., Urushihara H., Hernandez J., Rabbinowitsch E., Steffen D., Sanders M., Ma J., Kohara Y., Sharp S., Simmonds M., Spiegler S., Tivey A., Sugano S., White B., Walker D., Woodward J., Winckler T., Tanaka Y., Shaulsky G., Schleicher M., Weinstock G., Rosenthal A., Cox E.C., Chisholm R.L., Gibbs R., Loomis W.F., Platzer M., Kay R.R., Williams J., Dear P.H., Noegel A.A., Barrell B., Kuspa A. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ku C., Martin W.F. A natural barrier to lateral gene transfer from prokaryotes to eukaryotes revealed from genomes: the 70% rule. BMC Biol. 2016;14:89. doi: 10.1186/s12915-016-0315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ku C., Nelson-sathi S., Roettger M., Sousa F.L., Lockhart P.J., Bryant D., Hazkani-covo E., Mcinerney J.O., Landan G., Martin W.F. Endosymbiotic origin and differential loss of eukaryotic genes. Nature. 2015;524:427–437. doi: 10.1038/nature14963. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material Fig S4 Full rectangular view of phylogenetic tree showed in Fig. 1. Bootstraps support values above 50 are showed on the nodes.
Supplementary material Table S1 – List of Genbank Accession Numbers (NCBI) of retrieved eukaryotic sequences that belong to the Ohr/OsmC family. For each sequence, some features such as length, taxonomy and the signal peptide prediction are also listed.