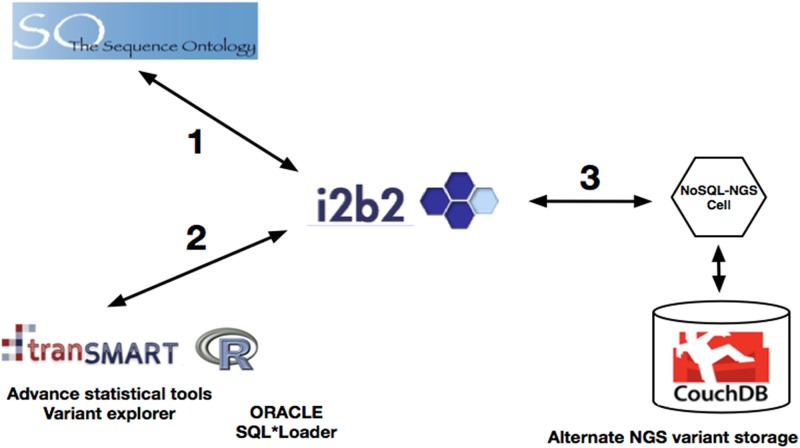
Fig 1. Overview of the three different approaches.
1) Using i2b2 by adding patient facts that have concepts coded per the Genome Sequence Ontology, 2) using i2b2/tranSMART by adding patient facts represented by a unique ontology allowing greater variant exploration, 3) using i2b2 by generating a patient set from i2b2 Star Schema database contained phenotypes and then using an alternate NoSQL-NGS variant storage to complete the genomic part of the query.