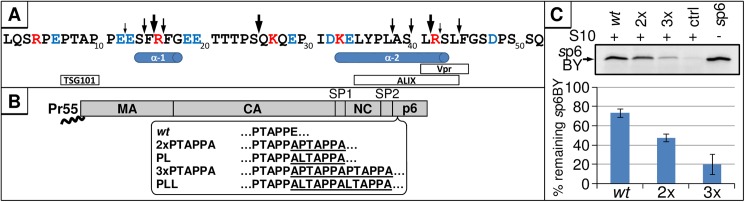
Fig 8. Generation of stable p6 mutants.
(A) 100 ng sp6 were incubated with 10 ng rIDE for 1, 5, 10, 30 or 60 min at 37°C. Reactions were stopped by adjusting the samples to 0.3% (w/v) TFE and subsequently analyzed by mass spectrometry. Arrows above the primary sequence represent the detected cleavage sites, and initial and secondary cleavage sites are indicated as big or small arrows, respectively. Red font indicates positively charged, and blue negatively charged residues. Previously identified α-helices and binding motifs are depicted below the primary sequence. (B) p6 mutants that encode multiple PTAPPA- or LTAPPA-motifs were generated. The introduced amino acids are underlined. (C) 10 ng of sp6BY were incubated with 250 μg/ml sp6 wt, 2xPTAPPA, 3xPTAPPA or only buffer and 5 μg S10 for 30 min at 37°C. sp6BY was detected by fluorescence emission and quantified. Values represent the arithmetic mean ± SD of four independent experiments.