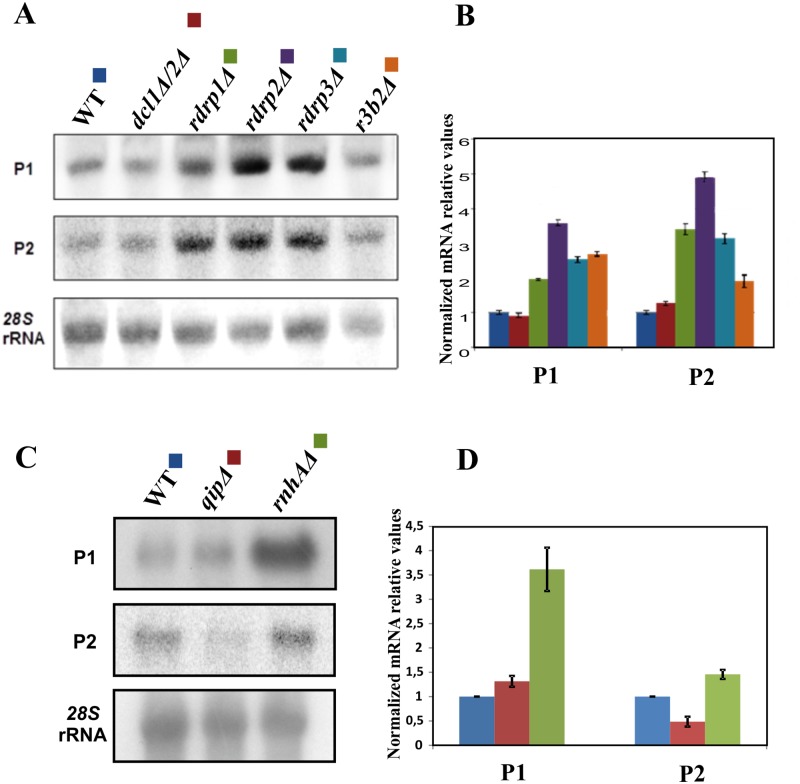
Fig 6. RdRP3 and RnhA participate in the rdrp-dependent dicer-independent RNA degradation pathway.
(A) Accumulation of mRNAs in wild type and silencing mutants. Northern blots of high molecular weight RNAs corresponding to genes regulated by the rdrp-dependent RNA degradation pathway (genes P1 and P2) were carried out using total RNA (50 μg) extracted from wild type (R7B), dcl1Δ/2Δ (MU411), r3b2Δ (MU412), rdrp1Δ (MU419), rdrp2Δ (MU420), and rdrp3Δ (MU438) mutant strains grown for 24 hours in liquid MMC medium. Samples were separated in 1.2% denaturing agarose gels, transferred to membranes, and hybridized with gene specific probes (S2 Table). Target genes correspond to the following gene products: P1: ID 26072, pyruvate decarboxylase, P2: ID 92956, actin binding protein. The membranes were reprobed with a 28S rRNA probe as loading control. Images are representative of three independent experiments. (C) Similarly, total RNA extracted from qip (MU430) and rnhA (MU437) mutant strains was analyzed in Northern blots assays. (B) and (D) Densitometric analysis of expression data shown in (A) and (C). Signal intensities were quantified and normalized to rRNA levels. All data were again normalized with respect to the expression value of the wild type strain (R7B) for each gene.