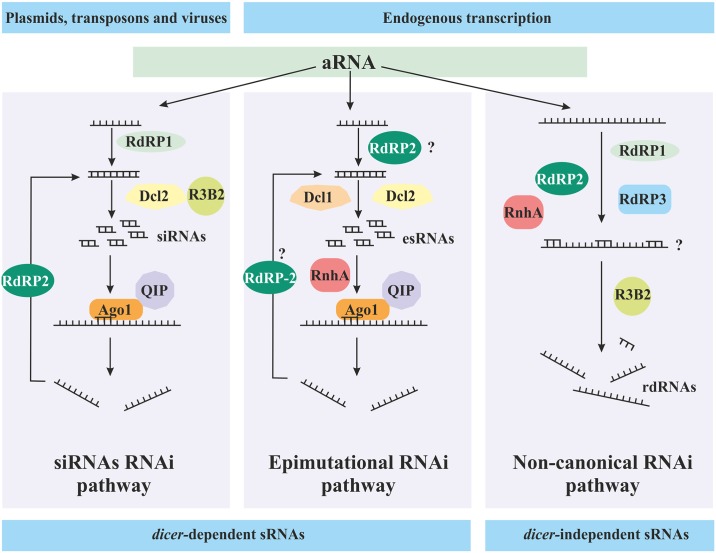
Fig 7. Models for the different RNAi pathways operating in M. circinelloides.
The siRNA RNAi pathway (left) is a defense mechanism against invasive nucleic acids such as plasmids, transposons, and viruses [36]. Aberrant transcripts from these invasive agents are used by RdRP1 to generate dsRNA molecules, which are cleaved by Dcl2 to produce siRNAs that are transferred to Ago1. The RNase III-like protein R3B2 participates in the biogenesis of these siRNAs, although its function in this pathway is still unknown. RdRP2 generates secondary dsRNA and amplifies this pathway [18]. The epimutation RNAi pathway (middle) silences target endogenous transcripts under stress conditions to generate epimutant strains that are better adapted. In this pathway, the generation of both primary and secondary dsRNA might be under the control of RdRP2, because rdrp2 mutants are incapable of producing epimutant strains resistant to the antifungal drug FK506 [4]. The function of RnhA might be mediated via its catalytic activity in unwinding ds-esRNAs; however, its precise role remains to be established. The non-canonical RNAi pathway (right) targets highly expressed mRNAs for degradation. RdRP1, RdRP2 [5] and RdRP3 (this study) could interact with these highly expressed mRNAs to synthesize complementary strands that signal these mRNAs for degradation by R3B2, which produces the resulting rdRNAs. RnhA (this study) has also a positive role in this pathway, although its specific function is not yet known.