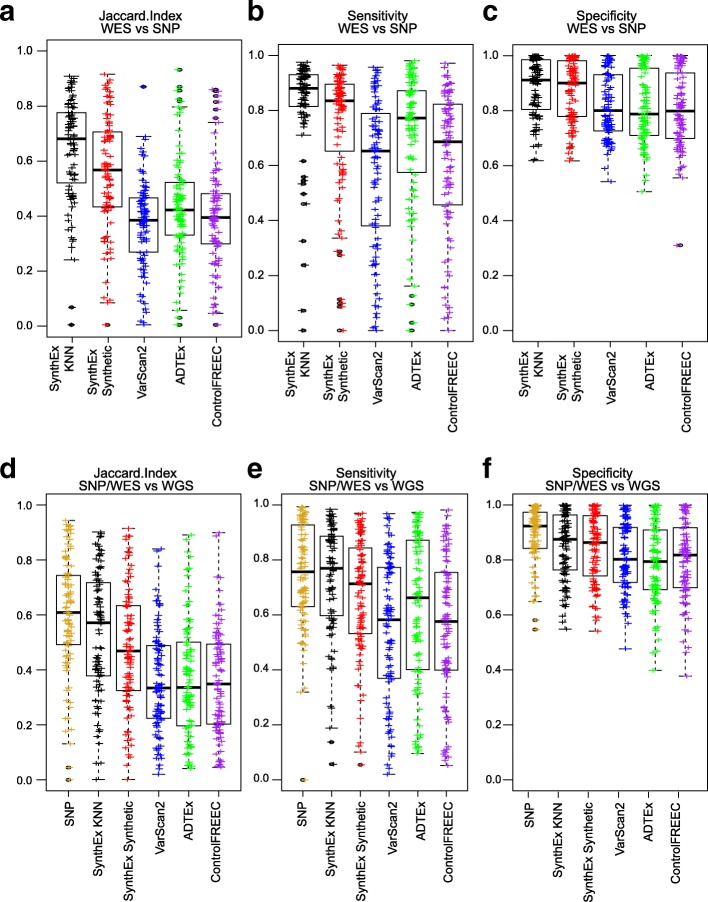
Fig. 7.
Jaccard Index, sensitivity, and specificity of SynthEx. Statistics of CNA tools from whole exome sequencing (WES) methods SynthEx KNN (black), SynthEx Synthetic (SN, red), VarScan2 (blue), ADTEx (green), and ControlFREEC (purple) compared to SNP arrays (a–c) and whole genome sequencing (WGS) (d–f) in TCGA breast cancer dataset. Compared to SNP arrays, one-sided t-tests tested SynthEx SN to the next highest mean to determine significance for a Jaccard Index (SynthEx SN versus ADTEx p = 4.9e-5), b sensitivity (SynthEx SN versus ADTEx p = 0.40), and c specificity (SynthEx SN versus VarScan2 p = 5.6e-10). Overall mean differences were significant in all plots as measured by ANOVA (Jaccard Index p = 1.7e-8, sensitivity p = 0.00029, specificity p = 0.0055). Compared to WGS, SNP (gold) and SynthEx KNN and SynthEx SN outperformed the other WES callers: d Jaccard Index (ANOVA p < 2e-16), e sensitivity (ANOVA p = 5.4e-9), and f specificity (ANOVA p = 1.5e-8). No statistical differences between SNP and SynthEx SN were measured in d–f (one-sided t-test p > 0.94)