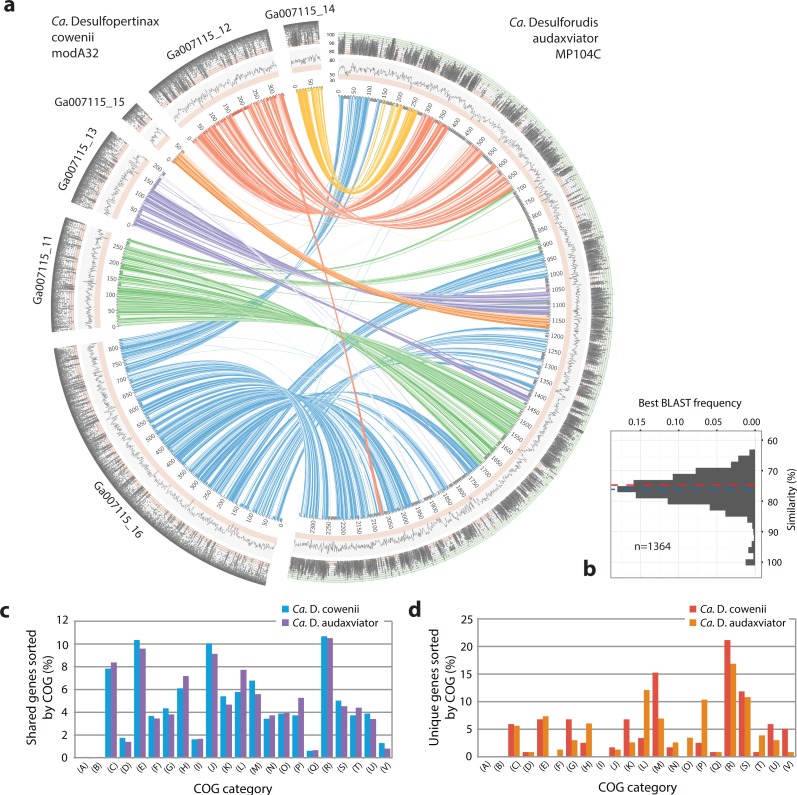
Figure 3. Analysis of genome alignment and shared and unique gene inventories in “Ca. Desulfopertinax cowenii” and “Ca. Desulforudis audaxviator.”.
Multiple genome alignment and analysis of shared and unique gene inventories reveal key conserved and variable features of “Ca. Desulfopertinax cowenii” and “Ca. Desulforudis audaxviator.” (A) Comparison of the “Ca. D. cowenii” genome scaffolds with “Ca. D. audaxviator” based on reciprocal best BLAST. From innermost to outermost, concentric circles show: nucleotide positions of genomes and scaffolds, percent GC content using a 100 bp sliding window, similarity of mapped U1362A reads. Links connecting circles are colored according to “Ca. D. cowenii” scaffold origin [Ga007115_(11–16)] and the degree of shading represents similarities (minimum similarity 70%) based on BLAST comparisons using <75% (light shade), ≥75% (dark shade) nucleic acid identity thresholds. (B) Frequency of reciprocal best BLAST hits (n = 1, 364) by percent similarity. Percent similarity histogram bins are in 2% increments and the dashed lines indicate average nucleotide identity (red) and average amino acid identity (blue) between “Ca. D. cowenii” and “Ca. D. audaxviator.” Relative abundance of shared (C) and unique (D) genes in the “Ca. D. cowenii” and “Ca. D. audaxviator” genomes, sorted by annotated COG categories. COG categories are: (A) RNA processing and modification; (B) Chromatin structure and dynamics; (C) Energy production and conversion; (D) Cell cycle control, cell division, chromosome partitioning; (E) Amino acid transport and metabolism; (F) Nucleotide transport and metabolism; (G) Carbohydrate transport and metabolism; (H) Coenzyme transport and metabolism; (I) Lipid transport and metabolism; (J) Translation, ribosomal structure and biogenesis; (K) Transcription; (L) Replication, recombination and repair; (M) Cell wall/membrane/envelope biogenesis; (N) Cell motility; (O) Post-translational modification, protein turnover, and chaperones; (P) Inorganic ion transport and metabolism; (Q) Secondary metabolites biosynthesis, transport, and catabolism; (R) General function prediction only; (S) Function unknown; (T) Signal transduction mechanisms; (U) Intracellular trafficking, secretion, and vesicular transport; (V) Defense mechanisms.