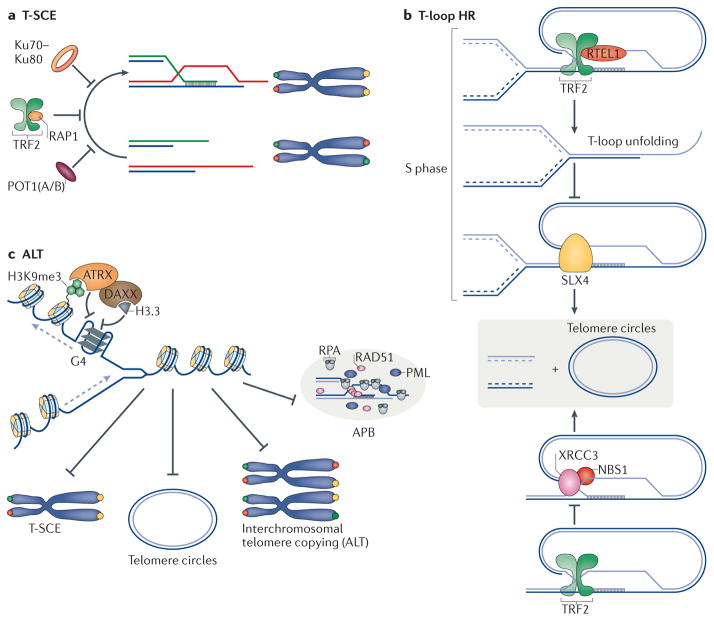
Figure 3. The three facets of telomere homologous recombination: T-SCE (telomere sister chromatid exchange), t-loop (telomere loop) homologous recombination and ALT (alternative lengthening of telomeres).
a | Exchange of sequence between sister chromatid telomeres (marked in red and green) is inhibited by RAP1 (repressor activator protein 1), POT1 (protection of telomere 1) and Ku70–Ku80. b | T-loop homologous recombination is blocked by TRF2 (telomere repeat-binding factor 2). TRF2 recruits RTEL1 (regulator of telomere elongation helicase 1) during S phase to unwind the t-loop and therefore protect it from being cleaved by structure-specific endonuclease subunit SLX4. In addition, TRF2 inhibits t-loop excision by inhibiting the activity of NBS1–XRCC3 (Nijmegen breakage syndrome 1–X-ray repair complementing defective repair in Chinese hamster cells 3). c | Telomere repeats have the propensity to form stable quadruplex (G4) DNA structures, which would impede replication fork progression. It has been proposed that ATRX (α-thalassaemia/mental retardation syndrome X-linked) unwind G4 DNA, enabling the deposition of histone H3.3 and ultimately assisting replication fork progression. The activity of ATRX at telomeres inhibits various ALT (alternative lengthening of telomeres) phenotypes including T-SCEs, formation of telomere circles, intrachromosomal telomere recombination and formation of APBs (ALT-associated promyelocytic leukaemia nuclear bodies). DAXX, death domain-associated protein; HR, homologous recombination; PML, promyelocytic leukaemia; RPA, replication protein A.