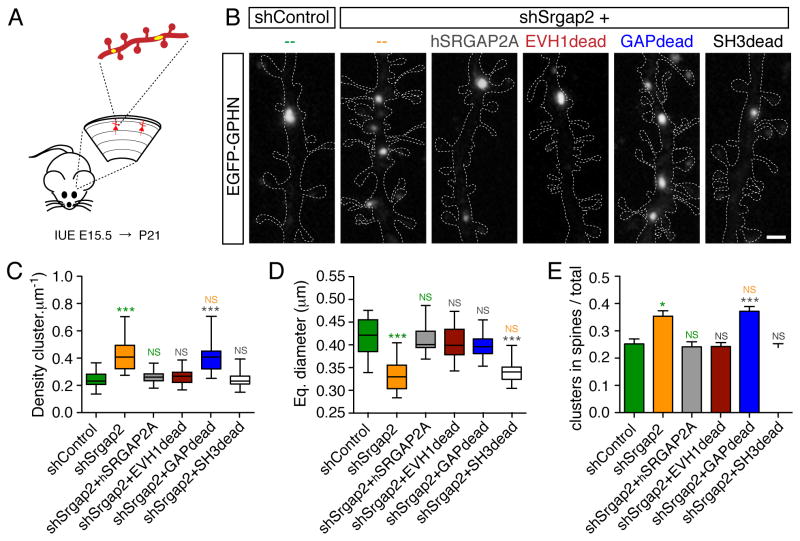
Figure 4. Molecular dissection of SRGAP2A function in inhibitory synaptic development in vivo.
(A) Schematic: segments of oblique dendrites of layer 2/3 cortical pyramidal neurons were imaged from P21 mice following sparse IUE. EGFP-Gephyrin (yellow) labels inhibitory synapses, TdTomato (red) allows the visualization of dendritic morphology.
(B) EGFP-Gephyrin clusters in representative segments of dendrites in control condition (shControl) or after in utero replacement of mouse Srgap2a with indicated mutants of hSRGAP2A. For clarity, dashed lines define the contours of TdTomato fluorescence in dendrites. Scale bar: 1 μm.
(C–E) Quantification of Gephyrin cluster density (C), Gephyrin cluster equivalent (Eq.) diameter (D) and mean proportion of Gephyrin clusters located in dendritic spines (± SEM) (E). nshControl = 32, nshSrgap2 = 36, nshSrgap2+hSRGAP2A = 31, nshSrgap2+EVH1dead = 30, nshSrgap2+GAPdead = 32, nshSrgap2+SH3dead = 31. *** p < 0.001, * p < 0.05, NS: p > 0.05, Kruskal-Wallis test followed by Dunn’s Multiple comparison test. Green, Orange, grey: comparison with shControl, shSrgap2 and shSrgap2+hSRGAP2A, respectively. See also Figure S6