Abstract
One of the human immunodeficiency virus (HIV) envelope proteins, gp41, plays a key role in HIV fusion. A gp41-derived peptide, T-20, efficiently inhibits HIV fusion and is currently approved for treatment of HIV-infected individuals. Although resistant variants have been reported, the mechanism of the resistance remains to be defined. To elucidate the mechanism in detail, we generated variants resistant to C34, a peptide derived from the gp41 carboxyl terminus heptad repeat (C-HR) in vitro. The resistant variants had a 5-amino-acid deletion in gp120 and a total of seven amino acid substitutions in gp41. Binding assays revealed that an I37K substitution in the N-terminal heptad repeat (N-HR) impaired the binding of C34, whereas an N126K substitution in the C-HR enhanced the binding to mutated N-HR, indicating that both mutations were directly involved in resistance. On the other hand, substitutions for A30 and D36 seemed to be secondary mutations, located complementary to each other in the Rev-responsive element (RRE), and were mutated simultaneously to maintain the secondary structure of the RRE that was impaired by the mutations at I37. Thus, HIV acquired resistance to C34 by mutations in N-HR, which directly interacted with C34. However, since this region also encoded the RRE, additional mutations were required to maintain viral replication. These results suggest that HIV fusion is one of the attractive targets for HIV chemotherapy.
Peptide inhibitors that block human immunodeficiency virus type 1 (HIV-1) fusion were first reported by Wild et al. (30). Recently, a peptide fusion inhibitor (T-20 or enfuvirtide) has been approved in the United States and Europe for treatment of HIV-infected individuals. The peptide sequence of T-20 is derived from the gp41 C terminus heptad repeat (C-HR) sequence, which corresponds to a linear region of 36 amino acids, and T-20 inhibits fusion by binding to the N-terminal heptad repeat (N-HR) of gp41 and preventing 6-helix bundle formation (4, 30). In HIV-infected patients, the effect of T-20 in combination with an antiretroviral regimen that was optimized with the aid of phenotypic and genotypic resistance testing (TORO 1 and 2) has been reported to suppress drug-resistant HIV replication more efficiently that the optimized regimen alone (17, 18).
The emergence of T-20-resistant HIV-1 was first reported in clinical patients receiving T-20 monotherapy in a phase I clinical trial (28) and subsequently in combined regimens employed in phase II and III trials of T-20 (23, 25). The T-20 susceptibility of recombinant HIV-1 containing the identified substitutions was examined in vitro and considered to be moderately resistant (5.4- to 6.3-fold) (28). However, the detailed mechanism of resistance of these variants still remains to be elucidated. On the other hand, Rimsky et al. revealed that three continuous amino acids in the N-HR (GIV at positions 36 to 38 of gp41) were crucial for the inhibition of HIV-1 entry by T-20 and for efficient association between N-HR and T-20 in vitro (26). Fikkert et al. also reported that HIV-1 variants resistant to T-20 contained substitutions in gp41, L33S and N43K, and a deletion of 5 amino acids, FNSTW (ΔFNSTW), in the V4 region of gp120 (9). L33S and N43K contributed to T-20 resistance, whereas the 5-amino-acid deletion alone had little effect on T-20 sensitivity. These results suggest that substitutions in the N-HR directly affect T-20 binding. Although the baseline sensitivity of HIV-1 to T-20 is defined by amino acid substitutions in gp41, coreceptor specificity is influenced by substitutions in the V3 loop in gp120, affects the fusion kinetics, and modulates T-20 sensitivity (4, 5).
To elucidate the mechanism of resistance to the peptide fusion inhibitors, we generated and characterized HIV-1 variants resistant to C34, a gp41 C-HR-derived peptide (2, 22) (Fig. 1A). During the selection of C34-resistant variants, we observed a 5-amino-acid deletion in the gp120 V4 region and a total of seven amino acid substitutions in gp41. Among the deletion and the substitutions, I37K and N126K play a key role in the resistance to C-HR-derived peptides, including T-20. Other deletions or substitutions were considered to enhance C34 resistance and/or improve the impaired replication kinetics. A30V and D36G maintained the Rev-responsive element (RRE) structure destabilized by I37T and I37K, respectively. Thus, these results reveal that the deletions or substitutions conferring resistance are restricted by both gp41 and RRE functions, suggesting that HIV-1 fusion is one of the most ideal targets for chemotherapy.
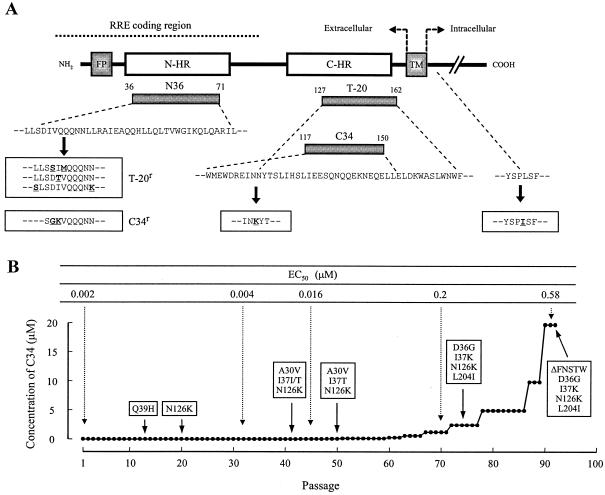
FIG. 1.
Schematic view of HIV-1 gp41 (A) and induction of C34-resistant HIV-1 (B). The locations of the fusion peptide (FP), N-terminal heptad repeat region (N-HR), C-terminal heptad repeat region (C-HR), transmembrane domain (TM), various gp41-derived peptides, and the Rev-responsive element (RRE) coding region are shown (A). The residue numbers of each peptide correspond to their positions in gp41. The bold underlined letters in the boxes indicate the novel mutations that have been reported in T-20-resistant HIV-1 variants (T-20r) in vitro (9, 26) and that have been observed in C34-resistant HIV-1 variants (C34r). (B) HIV-1NL4-3 was passaged in the presence of increasing concentrations of C34 in MT-2 cells. The dose-escalating selection was carried out for a total of 93 passages, with compound concentrations ranging from 0.0001 to 20 μΜ. At the indicated passages, proviral DNAs from the lysates of infected cells were sequenced, and the EC50s of the HIV-1 variants were determined with the MAGI assay.
MATERIALS AND METHODS
Cells and viruses.
MT-2 and Cos-7 cells were grown in RPMI 1640- and Dulbecco's modified Eagle medium-based culture medium, respectively. HeLa-CD4-LTR-β-gal cells were kindly provided by M. Emerman through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Disease (Bethesda, Md.), and used for the drug susceptibility assay (multinuclear activation of galactosidase indicator [MAGI] assay) as described previously (12, 14, 21). An HIV-1 infectious clone, pNL4-3, which was kindly provided by H. Sakai, Institute for Virus Research, Kyoto University (Kyoto, Japan), was used for constructions and the production of HIV-1 variants. A wild-type HIV-1, HIV-1WT, was generated by transfection of pNL4-3 into Cos-7 cells.
Antiviral agents.
The peptides used were N36, derived from the N-HR of gp41, and C34 and T-20, derived from the C-HR of gp41. The peptides were synthesized as described previously (24) and are depicted in Fig. 1A. 2′,3′-Dideoxycytidine (ddC) was purchased from Sigma (St. Louis, Mo.).
Determination of drug susceptibility of HIV-1.
The peptide sensitivity of infectious clones was determined by the MAGI assay with some modifications (14, 21). Briefly, the target cells (HeLa-CD4-LTR-β-gal; 104 cells/well) were plated in 96-well flat microtiter culture plates. On the following day, the cells were inoculated with the HIV-1 clones (60 MAGI U/well, giving 60 blue cells after 48 h of incubation) and cultured in the presence of various concentrations of drugs in fresh medium. Forty-eight hours after viral exposure, all the blue cells stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) were counted in each well. The activity of test compounds was determined as the concentration that blocked HIV-1 replication by 50% (50% effective concentration [EC50]).
Construction of recombinant HIV-1 clones.
Recombinant infectious HIV-1 clones carrying various mutations in gp120 and/or gp41 were generated by using pNL4-3. Briefly, the desired mutations were introduced into the NheI-BamHI region (1,220 bp) of pSLgp41WT, which encoded nucleotides 7250 to 8469 of pNL4-3, by an oligonucleotide-based mutagenesis method (29). NheI-BamHI fragments were inserted into pNL4-3, generating various molecular clones with the desired mutations. Each molecular clone was transfected into Cos-7 cells (105 cells/six-well culture plate). After 48 h, MT-2 cells (106 cells/well) were added and cocultured with the Cos-7 cells for an additional 24 h. When an extensive cytopathic effect (CPE) was observed, the supernatants were harvested and stored at −80°C until use.
Generation of HIV-1 variants resistant to C34.
MT-2 cells were exposed to HIV-1WT and cultured in the presence of C34 at an initial concentration of 0.0001 μM. Cultures were incubated at 37°C until an extensive CPE was observed. The culture supernatants were used for further passages in MT-2 cells in the presence of twofold increasing concentrations of C34 when massive CPEs were seen in the earlier periods. Such dose-escalating culture was performed until resistant variants were obtained. This selection was carried out for a total of 93 passages. At the indicated passages (Fig. 1B), the sequence of the env region was determined by direct sequencing of the proviral DNA extracted from the infected MT-2 cells.
Viral replication kinetics assay.
MT-2 cells (105 cells/5 ml) were infected with each virus preparation (500 MAGI U) for 4 h. The infected cells were then washed and cultured in a final volume of 5 ml. The culture supernatants (100 μl) were harvested on days 1, 2, 4, 6, and 8 after infection, and the p24 antigen amounts were determined.
For competitive HIV-1 replication assays (CHRA), two titrated infectious clones to be examined were mixed and added to MT-2 cells (105 cells/3 ml) as described previously (15) with some modifications. To ensure that the two infectious clones being compared were of approximately equal infectivity, a fixed amount (500 MAGI U) of one infectious clone was mixed with three different amounts (250, 500, and 1,000 MAGI U) of the other infectious clone. On day 1, one third of the infected MT-2 cells were harvested and washed twice with phosphate-buffered saline, and the cellular DNA was extracted. The purified DNA was subjected to nested PCR and then direct DNA sequencing. The HIV-1 coculture which best approximated a 50:50 mixture on day 1 was further propagated. Every 6 to 7 days, the cell-free supernatant of the virus coculture (1 ml) was transmitted to new uninfected MT-2 cells. The cells harvested at the end of each passage were subjected to direct sequencing, and the viral population change was determined.
Binding assay.
Each peptide (40 μM) was mixed with 10 mM phosphate-buffered saline-140 mM NaCl, pH 7.4, in an Aviv model 202 DS spectrometer equipped with a thermoelectric temperature controller. The thermal stability was assessed by monitoring the change in the circular dichroism signal at 222 nm. The midpoint of the thermal unfolding transition (melting temperature [Tm]) of each complex was determined as described previously (24).
Gel shift assay.
RNA of the RRE region and recombinant Rev were prepared as described previously (10) with some modifications. Briefly, the RRE region of the variants (nucleotides 7748 to 8009 of pNL4-3) was introduced into pBlueScript (Stratagene, La Jolla, Calif.). In vitro RNA transcription was performed with T7 RNA polymerase and [32P]UTP. Recombinant Rev was generated by use of the pGEX-6P-1/BL21 expression system (Amersham Biosciences, Piscataway, N.J.). The RNA and Rev were mixed at 25°C for 20 min in binding buffer (50 mM Tris-HCl, pH 7.5, 150 mM KCl, 1 mM dithiothreitol, 8% glycerol, 50 μg of tRNA/ml, and 100 μg of bovine serum albumin/ml) and subjected to native acrylamide gel electrophoresis.
RESULTS
Amino acid substitutions identified in the env region of C34-resistant HIV-1.
At passage 14 (P-14) in the culture where HIV-1 was propagating in the presence of C34 (0.0032 μM), one amino acid substitution, glutamine to histidine at position 39 (Q39H), in the N-HR of gp41 was transiently identified (Fig. 1B). At P-20 (0.0064 μM), a substitution, N126K, was newly identified in the C-HR, whereas Q39H had returned to the original wild-type amino acid. At P-41 (0.026 μM), two substitutions, A30V and I37I/T (mixture of I and T), were observed in the N-HR in addition to N126K, while at P-50 (0.077 μM), definitive I37T was detected (A30V/I37T/N126K) (Fig. 1B). At P-75 (2.5 μM), A30V had returned to the original wild-type amino acid, D36G was detected, I37T was substituted for I37K, and L204I, which was located in the cytoplasmic domain of gp41, was identified (D36G/I37K/N126K/L204I). At P-92 (20 μM), a deletion of five amino acids, FNSTW, in the V4 loop of gp120 (ΔFNSTW) was observed together with the four substitutions (ΔFNSTW/D36G/I37K/N126K/L204I) (Fig. 1B). In addition to the env region, we also examined the Tat- and Rev-encoding regions but did not observe any substitutions. These results suggest that, in order to develop a higher resistance to C34, HIV-1 acquires not only multiple substitutions in gp41 but also the 5-amino-acid deletion in gp120.
Susceptibility of the different env recombinant viruses to fusion inhibitors.
To clarify which substitutions among the identified changes were responsible for C34 resistance, we first generated infectious HIV-1 clones containing the deletion (ΔFNSTW) in gp120 or the single amino acid substitutions (A30V, D36G, I37T, I37K, Q39H, N126K, or L204I) in gp41 that were observed during the selection procedure (Fig. 1B). We also evaluated the activities of the gp41-derived peptides N36, T-20, and C34 and a reverse transcriptase inhibitor used as a control, ddC, against these strains with the MAGI assay (Table 1).
TABLE 1.
Antiviral activity of HIV-1 gp41-derived peptides against gp120 and/or gp41 recombinant virusesa
| Virus or substitution | EC50 (nM)
|
|||
|---|---|---|---|---|
| ddC | T-20 | N36 | C34 | |
| HIV-1WTb | 264 | 12 | 51 | 2.1 |
| ΔFNSTWc | 98 (0.4) | 18 (1.5) | 50 (1.0) | 9.5 (4.6) |
| A30V | 205 (0.8) | 6.3 (0.5) | 38 (0.7) | 7.0 (3.4) |
| D36G | 173 (0.7) | 0.92 (0.1) | 90 (1.7) | 1.6 (0.8) |
| D36Se | 166 (0.6) | 6.6 (0.6) | 89 (1.7) | 3.7 (0.6) |
| I37T | 284 (1.1) | 156 (13) | 40 (0.8) | 23 (11) |
| I37K | 326 (1.2) | 2,482 (212) | 99 (1.9) | 27 (13) |
| V38Me | 223 (0.8) | 305 (26) | 94 (1.8) | 31 (15) |
| Q39Hd | 330 (1.3) | 1.9 (0.2) | 165 (3.2) | 5.3 (2.6) |
| N126Kd | 380 (1.4) | 23 (1.9) | 137 (2.7) | 14 (6.8) |
| L204I | 247 (0.9) | 13 (1.1) | 105 (2.0) | 4.4 (2.1) |
| D36S/V38Me | 294 (1.1) | 60 (5.1) | 46 (0.9) | 16 (7.7) |
| I37T/N126K | 292 (1.1) | 158 (14) | 54 (1.1) | 22 (11) |
| I37K/N126K | 309 (1.2) | 1,570 (134) | 51 (1.0) | 57 (28) |
| A30V/I37K/N126K | 409 (1.5) | 198 (17) | 119 (2.3) | 22 (10) |
| D36G/I37K/N126K | 329 (1.2) | 269 (23) | 156 (3.0) | 148 (72) |
| D36G/I37K/N126K/L204Id | 209 (0.8) | 117 (10) | 41 (1.2) | 112 (54) |
| ΔFNSTW/D36G/I37K/N126K/L204Id | 213 (0.8) | 746 (64) | 54 (1.0) | 171 (83) |
Anti-HIV activity was determined with the MAGI assay. The data shown are mean values obtained from the results of at least three independent experiments, and resistance (n-fold) in EC50 for recombinant viruses compared to HIV-1WT is shown in parentheses.
HIV-1NL4-3 was used was a wild-type virus.
ΔFNSTW is the deletion of 5 amino acids at positions 364 to 368 in the gp120 V4 region of HIV-1NL4-3.
Mutant viruses observed during induction of C34 resistance variants in vitro (Fig. 1B).
D36S/V38M has been reported for T-20-resistant HIV-1 variants (26).
HIV-1ΔFNSTW, HIV-1A30V, HIV-1Q39H, and HIV-1L204I showed weak resistance to C34 compared with HIV-1WT (less than fivefold). Interestingly, D36G, observed in the majority of HIV-1 strains (16), conferred an increased T-20 susceptibility to HIV-1 (10-fold), in agreement with previous reports (20, 26), whereas D36G did not contribute C34 resistance by itself (0.8-fold). Although I37T has also been reported as one of the T-20 resistance mutations in vitro, its detailed mechanism of resistance remains unknown (20, 26). In our experiments, I37T conferred T-20 and C34 resistance to HIV-1 (13- and 11-fold, respectively), and I38K also conferred both T-20 and C34 resistance (212- and 13-fold, respectively). HIV-1N126K showed moderate resistance to C34 (6.8-fold). Neither the deletion in gp120 nor any of the substitutions in gp41 conferred resistance to N36 or ddC (Table 1).
Although the I37 substitutions appeared to be primarily responsible for C34 resistance, the C34 resistance levels of the I37 substitution variants were not comparable to that of the selected virus at P-93 (EC50, 0.78 μM). Therefore, we generated infectious HIV-1 clones containing the identified substitutions combined with I37T or I37K and determined their susceptibilities to the peptides (Table 1). The combination of I37K and N126K enhanced C34 resistance (13- to 28-fold), whereas HIV-1I37T, HIV-1I37T/N126K, and HIV-1A30V/I37T/N126K showed comparable resistance levels to C34. Moreover, I37K/N126K combined with D36G (D36G/I37K/N126K) enhanced C34 resistance (72-fold), although the L204I substitution combined with D36G/I37K/N126K decreased the levels of resistance to both T-20 and C34 (10- and 54-fold, respectively). A clone containing the deletion in gp120 and four substitutions in gp41, HIV-1ΔFNSTW/D36G/I37K/N126K/L204I, showed the highest resistance to C34 (83-fold) and cross-resistance to T-20 (64-fold). These results indicate that the I37K substitution is mainly responsible for C34 resistance, whereas the other substitutions enhance the resistance or improve the impaired viral replication kinetics.
Next, we generated a T-20-resistant molecular clone which had been previously reported (26), HIV-1D36S/V38M, and evaluated the susceptibility to N36, T-20, and C34. HIV-1D36S/V38M showed moderate resistance to both T-20 and C34 (5.1- and 7.7-fold, respectively) (Table 1). We also generated HIV-1 variants that contained each of the single substitutions, HIV-1D36S and HIV-1V38M. HIV-1D36S did not contribute to the resistance, although HIV-1V38M showed cross-resistance to T-20 and C34 (26- and 15-fold, respectively). Combined with the finding that I37K is the major mutation for resistance to C34, this region, positions 37 and 38 of gp41, appears to be involved in resistance to both T-20 and C34, while changes at position 36 appear to be largely restricted in their effects to T-20.
Peptide binding affinity.
To clarify the effect of the substitutions on the interaction of N-HR and C-HR, the binding affinity of the peptides in vitro was examined with the synthesized peptides (Table 2). The affinity between N36D36G/I37K and C34 was unstable even at 37°C, indicating that the peptide inhibitor C34 hardly bound to N36D36G/I37K. However, it is still unclear whether it is a direct effect of the N-HR mutations decreasing the affinity of C34 binding or an indirect effect of the N-HR mutations destabilizing the N-HR trimer formation. In contrast, C34N126K, with the substitution responsible for the resistance, showed enhanced binding affinity not only to N36 but also to N36D36G/I37K. Thus, there are two implications of mutations in gp41 for conferring C34 resistance: the decreased affinity of C34 for N36D36G/I37K and the increased affinity of C34N126K for both N36WT and N36D36G/I37K. In other words, the D36G and I37K substitutions in the N-HR interfere with the binding of the peptide inhibitors, such as T-20 and C34, and N126K in the C-HR enhances the intra-gp41 binding of N-HR and C-HR compared with the peptide inhibitors.
TABLE 2.
Binding affinity of wild and mutated peptidesa
| Peptide | Tm (°C) |
|---|---|
| N36/C34 | 49.5 |
| N36D36G/I37K/C34 | 38.5 |
| N36/C34N126K | 55.0 |
| N36D36G/I37K/C34N126K | 45.0 |
The binding affinity of the peptides is shown with Tm values.
Replication kinetics of C34-resistant variants.
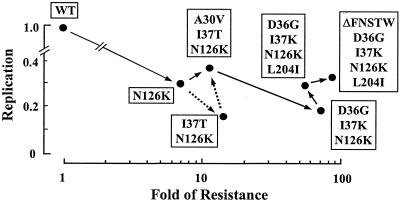
To determine the effects of the identified deletion and mutations on HIV-1 replication, we first examined the replication kinetics of HIV-1 variants by p24 production in the culture supernatants. The p24 production by the variants ranged from 14 to 34% of that of HIV-1WT (HIV-1I37T/N126K, 14%; HIV-1N126K, 30%; and HIV-1A30V/I37T/N126K, 34%) as determined at day 8 (Fig. 2). Next, the replication levels of variants with representative substitutions were compared by CHRA. The resistances (n-fold) of the variants are also shown in Fig. 2. Since HIV-1Q39H was considered to be one of the polymorphisms and appeared only transiently, we first compared the replication of HIV-1WT and HIV-1N126K and found an impaired replication profile for HIV-1N126K. The variant with a combination including the I37T substitution, HIV-1I37T/N126K, which was not observed during selection, showed the slowest replication profile. To develop an HIV-1A30V/I37T/N126K variant, the A30V substitution was introduced first, and then the I37T substitution was introduced (Fig. 1B). This was consistent with the results of the CHRA that the replication profile of HIV-1A30V/I37T/N126K was greater than that of HIV-1I37T/N126K, suggesting that the A30V substitution improved the impaired replication profile of HIV-1I37T/N126K. Since both HIV-1I37K and HIV-1I37K/N126K could replicate for only a few passages, these variants could not be studied. For I37K/N126K substitutions, D36G instead of A30V substitution seemed to be suitable in order to maintain resistance and replication, although the replication of HIV-1D36G/I37K/N126K still remained impaired. The L204I substitution improved the replication kinetics of HIV-1D36G/I37K/N126K. The deletion in the V4 region contributed to both the increased resistance and replication. These results indicate that among the deletion and the substitutions, I37T/K and N126K, and especially I37K, are related to resistance, while A30V, D36G, and L204I are associated with improvement of replication and the deletion in the V4 region had modest effects on increased resistance and replication.
FIG. 2.
Replication kinetics of the resistant variants. The replication kinetics determined by p24 antigen production and the CHRA are summarized. The data are depicted as the resistance (x axis) and replication (y axis) compared with those of HIV-1WT. Variants observed (continuous arrows) and not observed (dashed arrows) in the selection are shown in the order of their emergence.
Rev and RRE interactions.
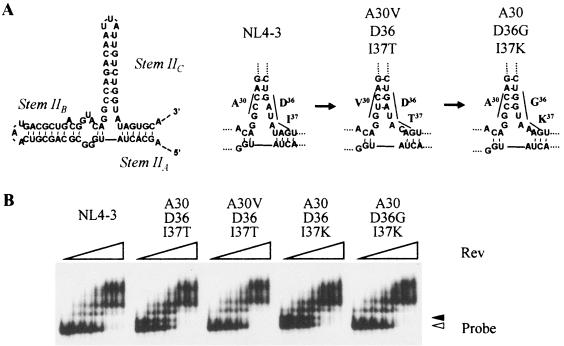
The nucleotides for the substitutions A30V, D36G, and I38T/K also encode the RRE, and the nucleotides for A30 and D36 are located complementary to each other in stem IIC (Fig. 3A). A30V emerged along with I37T and intact D36, and D36G was found with I37K and intact A30. Thus, these mutations were thought to maintain the positioning of stems IIA and IIB in the tertiary structure of the RRE. To examine the effect of these nucleotide substitutions on the Rev-RRE interaction, we performed a gel shift assay. The secondary structure of the RRE that contained the I37T or I37K substitutions showed two forms (Fig. 3B). Introduction of A30V, which could improve the replication of HIV-1I37T, changed the two RRE structures into a structure identical to that observed in the wild type. D36G observed with I37K also changed the separated RRE structure. However, little difference (less than 10%) in the Rev binding caused by the substitutions was observed between the wild-type and the mutated RRE (Fig. 3B). These results indicate that substitutions at A30 or D36 maintain the stability of the RRE secondary structure that is affected by nucleotide substitutions at I37.
FIG. 3.
Putative secondary structure of the RRE and locations of the nucleotides corresponding to the substitutions. The IIA, IIB, and IIC stems and the effects of the nucleotide substitutions are shown (A). A30V (GCC to GTC) to D36 (GAT) and A30 (GCC) to D36G (GAT to GGT) are located complementary to each other in the stem IIB (underlined). The effects of the nucleotide substitutions on the Rev-RRE interaction were examined by gel shift assays (B). The RRE of HIV-1I37T and HIV-1I37K displayed two signals. The amounts of Rev used were 0, 20, 40, 80, 160, 240, and 320 nM (left to right).
DISCUSSION
After adsorption to cells followed by conformational changes of gp120, HIV fusion takes place by interaction and binding of the N-HR and C-HR of gp41. C-HR-derived peptides, e.g., T-20 or C34, inhibit HIV replication as a decoy for C-HR (2, 30). Previous reports showed that mutations in N-HR (19) were associated with resistance to T-20 (9, 25, 26, 28). In this study, we showed that competition between C34 and C-HR also plays a key role in acquiring resistance. However, in order to develop high resistance to C34, changes in the binding affinity of N-HR, as observed in T-20 resistance, are insufficient on their own, and an additional mutation in C-HR is necessary. According to the crystal structure of gp41 (2), none of the substitutions observed in this study are located on the binding surface of the N-HR or C-HR. Even one of the primary mutations that directly contribute to the resistance, I37K, is not located on the binding surface of N-HR. We also identified a primary mutation, N126K, that is located outside of the binding surface of C-HR. Although the significance of these locations still remains to be defined at present, further structural analysis may provide insights into the mechanism of binding enhancement. Our observations also provide evidence that small compounds interacting directly with the binding surface seem to inhibit HIV replication efficiently, since HIV would hardly be mutated on the binding surface.
During the selection of the C34-resistant variants, the substitutions were introduced in the following order: a substitution (N126K) associated with susceptibility to C34 was introduced first, followed by a substitution (A30V) associated with replication. L204I also improved the replication kinetics of HIV-1D36G/I37K/N126K. It is likely that substitutions that are associated with resistance usually impair the replication kinetics, resulting in selection of HIV-1 variants containing substitutions that improve the replication disadvantages. This hypothesis has been proved with analyses of replication kinetics of T-20-resistant variants described previously (20). Moreover, such substitution patterns have previously been observed in multi-dideoxynucleoside-resistant variants (15, 21). However, the mechanism of replication improvement in multi-dideoxynucleoside-resistant variants remains unknown. In this study, I37 is one of the key amino acids for C34 resistance, and it is located in an important region for the Rev-RRE interaction (13). The significance of the secondary mutations, A30V and D36G, for improvement of the RRE structural stability impaired by I37T or I37K is thought to be that they maintain both gp41 and RRE functions. In contrast to the C34 resistance mutations, nucleotides encoding some T-20 resistance mutations, L33S and N43K (9), are located in a single-stranded bulge region of the stem IIC loop top (UUA to UCA) (Fig. 3A) and in the bulge region of stem III, indicating that the structural changes to the RRE would be minimal, while other T-20 resistance mutations (20), such as G36D, G36S, I37K, and possibly V38A and V38 M, appear to alter the stability of the RRE stem IIC structure and to impair the replication kinetics. To date, the Rev-RRE interaction has mainly been examined for stem IIB, since Rev directly binds to it (10, 13). Although the functions of stems IIA and IIC remain to be defined, it is possible that the secondary mutations in stem IIC influence the Rev-RRE interaction, since we have shown that the secondary mutations were introduced simultaneously with the primary mutations and improved replication. These results also indicate that the conformation of the RRE is essential for the Rev-RRE interaction and not just the nucleotide sequence of the RRE itself.
NL4-3 gp41 contains four N-glycan attachment sites, N-X-S/T, located at N100-A-S, N105-K-S, N114-M-T, and N126-Y-T. These four sites are highly conserved in various HIV strains (11, 16). Mutational analysis revealed that each substitution of the N glycosylation sites had a modest effect on HIV replication, whereas some combined substitutions severely impaired replication (11). Although the effect of N126-glycan on binding of the N-HR and C-HR remains unknown, it would be possible that N126-glycan plays some roles for C34 resistance. Different effects of N126K substitution on susceptibility to T-20 and C34 (1.9- and 6.8-fold, respectively) were observed (Table 1). This result might be accounted for by the finding that N126 locates at −1 (outside) from the N terminus of T-20, whereas it locates inside (+10) of C34 (Fig. 1A).
It has been reported that a tyrosine-based sorting signal in the gp41 cytoplasmic domain, Y201-X-X-L, was involved in trafficking and targeting to the plasma membrane of the gp41 (3). The motif is highly conserved among various HIV strains (16, 19). Although the role of Y201 for infectivity has been studied in detail (3, 19), that of L204 remains to be defined. In the present study, we showed that the L204I substitution enhanced viral replication, suggesting that L204, as well as Y201, plays an important role for viral replication.
pNL4-3 was established as a molecular clone of wild-type HIV-1 (1) and is widely used in HIV research. However, it represents only one of the wild-type HIV-1 variants. In fact, even in the absence of C34, we still observed several substitutions in NL4-3 that were identified in the C34 selection, e.g., A30V and Q39H. These substitutions are also observed in some treatment-naïve clinical isolates (16). It is well known that HIV reverse transcriptase makes several nucleotides miss incorporation during the reverse transcription, suggesting that each HIV isolate, even in the wild-type population, contains several substitutions in the integrated DNA genome. D36 is identified only in pNL4-3-derived clones, although the G36/I37/V38 motif is well conserved, not only in HIV-1 but also in HIV-2 and simian immunodeficiency virus strains (16). Furthermore, the 5-amino-acid deletion in gp120 was reported not only in a fusion inhibitor, T-20 (9), but also in CD4-gp120-binding inhibitors DS5000 (8) and AR177 (Zintevir) (7), CXCR4 antagonists, bicyclams JM2763 and SID791 (6), and SDF-1α-resistant variants (27). In these reports, pNL4-3-derived viruses were also used for the selection of the resistant variants. Only pNL4-3 has the 5-amino-acid tandem sequence FNSTWFNSTW in the gp120 V4 region. Therefore, this deletion is thought to be specific for HIV-1NL4-3, although the 5-amino-acid deletion conferred weak C34 resistance. These results indicate that we should be careful before concluding that such substitutions are involved in the resistance or replication kinetics.
In conclusion, HIV acquires resistance against C34 by mutations in both N-HR and C-HR. However, mutations in N-HR are restricted by Rev-RRE and/or gp120-gp41 interactions, suggesting that HIV-1 fusion is one of the most attractive targets for blocking HIV infection.
Acknowledgments
We thank Hiroaki Mitsuya for helpful suggestions and Ayako Yoshioka for manuscript preparation.
This work was supported in part by a grant for the Promotion of AIDS Research from the Ministry of Health and Welfare of Japan (M.M.), a grant for Research for Health Sciences Focusing on Drug Innovation from the Japan Health Sciences Foundation (E.K.), and a grant from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (E.K.). D.N. is supported by the 21st Century COE Program of the Ministry of Education, Culture, Sports, Science, and Technology.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of the acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 3.Day, J. R., C. Munk, and J. C. Guatelli. 2004. The membrane-proximal tyosine-based sorting signal of human immunodeficiency virus type 1 gp41 is required for optimal viral infectivity. J. Virol. 78:1069-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, X. Wu, W. A. O'Brien, L. Ratner, J. C. Kappes, G. M. Shaw, and E. Hunter. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74:8358-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, Z. Zhang, W. A. O'Brien, L. Ratner, G. M. Shaw, and E. Hunter. 2001. Sensitivity of human immunodeficiency virus type 1 to fusion inhibitors targeted to the gp41 first heptad repeat involves distinct regions of gp41 and is consistently modulated by gp120 interactions with the coreceptor. J. Virol. 75:8605-8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Vreese, K., V. Kofler-Mongold, C. Leutgeb, V. Weber, K. Vermeire, S. Schacht, J. Anne, E. De Clercq, R. Datema, and G. Werner. 1996. The molecular target of bicyclams, potent inhibitors of human immunodeficiency virus replication. J. Virol. 70:689-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Este, J. A., C. Cabrera, D. Schols, P. Cherepanov, A. Gutierrez, M. Witvrouw, C. Pannecouque, Z. Debyser, R. F. Rando, B. Clotet, J. Desmyter, and E. De Clercq. 1998. Human immunodeficiency virus glycoprotein gp120 as the primary target for the antiviral action of AR177 (Zintevir). Mol. Pharmacol. 53:340-345. [DOI] [PubMed] [Google Scholar]
- 8.Este, J. A., D. Schols, K. De Vreese, K. Van Laethem, A. M. Vandamme, J. Desmyter, and E. De Clercq. 1997. Development of resistance of human immunodeficiency virus type 1 to dextran sulfate associated with the emergence of specific mutations in the envelope gp120 glycoprotein. Mol. Pharmacol. 52:98-104. [DOI] [PubMed] [Google Scholar]
- 9.Fikkert, V., P. Cherepanov, K. Van Laethem, A. Hantson, B. Van Remoortel, C. Pannecouque, E. De Clercq, Z. Debyser, A.-M. Vandamme, and M. Witvrouw. 2002. env chimeric virus technology for evaluating human immunodeficiency virus susceptibility to entry inhibitors. Antimicrob. Agents Chemother. 46:3954-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson, B. R. 1997. Interaction between HIV Rev and nuclear import and export factors: the Rev nuclear localisation signal mediates specific binding to human importin-β. J. Mol. Biol. 274:693-707. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, W. E., J. M. Sauvron, and R. C. Desrosiers. 2001. Conserved, N-linked carbohydrates of human immunodeficiency virus type 1 gp41 are largely dispensable for viral replication. J. Virol. 75:11426-11436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kjems, J., M. Brown, D. D. Chang, and P. A. Sharp. 1991. Structural analysis of the interaction between the human immunodeficiency virus Rev protein and the Rev response element. Proc. Natl. Acad. Sci. USA 88:683-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kodama, E. I., S. Kohgo, K. Kitano, H. Machida, H. Gatanaga, S. Shigeta, M. Matsuoka, H. Ohrui, and H. Mitsuya. 2001. 4′-Ethynyl nucleoside analogs: potent inhibitors of multidrug-resistant human immunodeficiency virus variants in vitro. Antimicrob. Agents Chemother. 45:1539-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosalaraksa, P., M. F. Kavlick, V. Maroun, R. Le, and H. Mitsuya. 1999. Comparative fitness of multi-dideoxynucleoside-resistant human immunodeficiency virus type 1 (HIV-1) in an in vitro competitive HIV-1 replication assay. J. Virol. 73:5356-5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuiken, C., B. Foley, B. Hahn, P. Marx, F. McCutchan, J. Mellors, S. Wolinsky, and B. Korber (ed.). 2001. HIV sequence compendium 2001. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.M.
- 17.Lalezari, J. P., K. Henry, M. O'Hearn, J. S. Montaner, P. J. Piliero, B. Trottier, S. Walmsley, C. Cohen, D. R. Kuritzkes, J. J. Eron, Jr., J. Chung, R. DeMasi, L. Donatacci, C. Drobnes, J. Delehanty, and M. Salgo. 2003. Enfuvirtide, an HIV-1 fusion inhibitor, for drug-resistant HIV infection in North and South America. N. Engl. J. Med. 348:2175-2185. [DOI] [PubMed] [Google Scholar]
- 18.Lazzarin, A., B. Clotet, D. Cooper, J. Reynes, K. Arasteh, M. Nelson, C. Katlama, H. J. Stellbrink, J. F. Delfraissy, J. Lange, L. Huson, R. DeMasi, C. Wat, J. Delehanty, C. Drobnes, and M. Salgo. 2003. Efficacy of enfuvirtide in patients infected with drug-resistant HIV-1 in Europe and Australia. N. Engl. J. Med. 348:2186-2195. [DOI] [PubMed] [Google Scholar]
- 19.Lodge, R., J.-P. Lalonde, G. Lemay, and E. A. Cohen. 1997. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J. 16:695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu, J., P. Sista, F. Giguel, M. Greenberg, and D. Kuritzkes. 2004. Relative replicative fitness of human immunodeficiency virus type 1 mutants resistant to enfuvirtide (T-20). J. Virol. 78:4628-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maeda, Y., D. J. Venzon, and H. Mitsuya. 1998. Altered drug sensitivity, fitness, and evolution of human immunodeficiency virus type 1 with pol gene mutations conferring multi-dideoxynucleoside resistance. J. Infect. Dis. 177:1207-1213. [DOI] [PubMed] [Google Scholar]
- 22.Malashkevich, V. N., D. C. Chan, C. T. Chutkowski, and P. S. Kim. 1998. Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: conserved helical interactions underlie the broad inhibitory activity of gp41 peptides. Proc. Natl. Acad. Sci. USA 95:9134-9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matthews, T., M. Salgo, M. Greenberg, J. Chung, R. DeMasi, and D. Bolognesi. 2004. Enfuvirtide: the first therapy to inhibit the entry of HIV-1 into host CD4 lymphocytes. Nat. Rev. Drug Discov. 3:215-225. [DOI] [PubMed] [Google Scholar]
- 24.Otaka, A., M. Nakamura, D. Nameki, E. Kodama, S. Uchiyama, S. Nakamura, H. Nakano, H. Tamamura, Y. Kobayashi, M. Matsuoka, and N. Fujii. 2002. Remodeling of gp41-C34 peptide leads to highly effective inhibitors of the fusion of HIV-1 with target cells. Angew. Chem. Int. Ed. Engl. 41:2937-2940. [DOI] [PubMed] [Google Scholar]
- 25.Poveda, E., B. Rodes, C. Toro, L. Martin-Carbonero, J. Gonzalez-Lahoz, and V. Soriano. 2002. Evolution of the gp41 env region in HIV-infected patients receiving T-20, a fusion inhibitor. AIDS 16:1959-1961. [DOI] [PubMed] [Google Scholar]
- 26.Rimsky, L. T., D. C. Shugars, and T. J. Matthews. 1998. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72:986-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schols, D., J. A. Este, C. Cabrera, and E. De Clercq. 1998. T-cell-line-tropic human immunodeficiency virus type 1 that is made resistant to stromal cell-derived factor 1α contains mutations in the envelope gp120 but does not show a switch in coreceptor use. J. Virol. 72:4032-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weiner, M. P., G. L. Costa, W. Schoettlin, J. Cline, E. Mathur, and J. C. Bauer. 1994. Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene 151:119-123. [DOI] [PubMed] [Google Scholar]
- 30.Wild, C., T. Oas, C. McDanal, D. Bolognesi, and T. Matthews. 1992. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc. Natl. Acad. Sci. USA 89:10537-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]