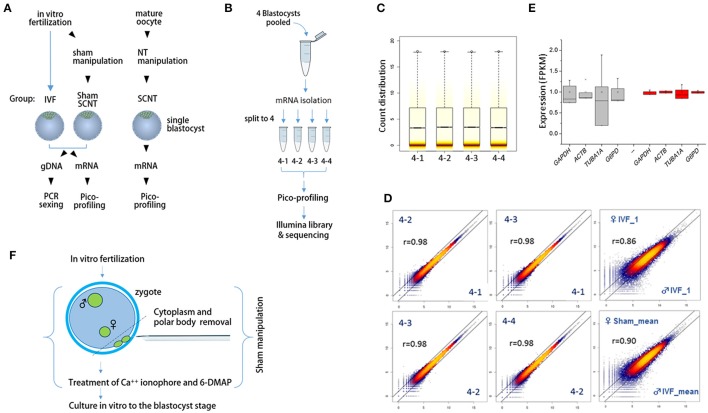
Figure 1.
Schematics of the sample preparation and pico-profiling processes for deep sequencing. (A) Preparation of bovine blastocyst samples for sexing and pico-profiling. Blastocysts were derived by in vitro-fertilization (IVF) or somatic cell nuclear transfer (SCNT). Adult ear skin fibroblasts were used as donor cells to obtain SCNT embryos. Genomic DNA and mRNA were extracted from single IVF and sham nuclear transfer (NT) blastocysts for sexing and pico-profiling. (B–E) Validation of the pico-profiling method. In (B), there is an overview of the experimental strategy for validation using four identical replicates of pooled blastocyst samples. In (C) and (D), sequencing results are presented. Box plot in (C) shows the distribution of total gene expression levels [log2(FPKM)] for each replicate. Colored stripes indicate gene density within a specific expression level. Scatter plots in (D) show the correlation between replicates. For comparison, other non-technical replicates (i.e., male IVF and female IVF blastocyst samples) are included. Pearson correlation coefficients (r) are denoted on each plot. In E, expression levels of housekeeping genes are compared between replicate groups (red) and other independent sample groups (gray). Note significantly less variability in the expression levels of the replicates compared to that found in other sample groups. (F), Generation of sham NT embryos. Sixteen to 18 h after IVF, a zygote with maternal and paternal pronuclei was manipulated to remove the polar bodies plus a portion of cytoplasm underneath them, followed by a standard nuclear transfer protocol to develop to the blastocyst stage.