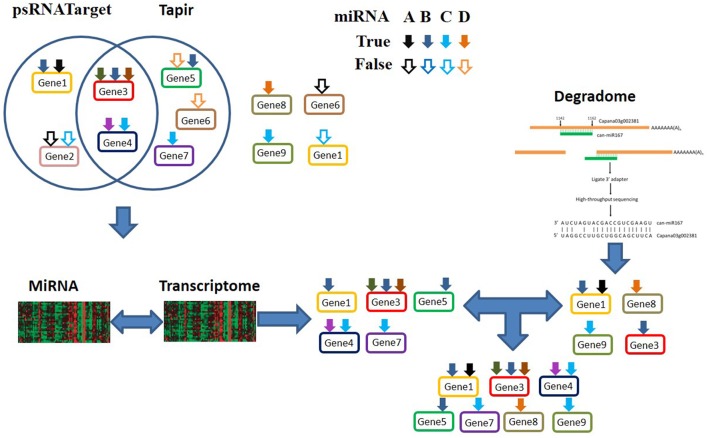
Figure 2.
An example for the miRNA targets prediction by MiRTrans. Seven genes (Gene1 to Gene7) are assumed to be putative target genes of four miRNAs: A–D (demonstrated as black, dark blue, light blue and orange arrows), according to the non-redundant combination of psRNATarget and Tapir predictions. False positive miRNA-transcript pairs: A to Gene2, C to Gene2, D to Gene5, D to Gene6 are included the in sequence-based predictions. D to Gene8 and C to Gene9 are incorrectly missing in sequence-based predictions. By identifying the dependency between the expression of miRNAs and transcripts, MiRTrans refines the predictions by removing the false miRNA-transcript pairs from sequence-based predictions. Degradome sequencing data are incorporated to recoup those targets falsely removed by the previous steps (A to Gene1, B to Gene3, D to Gene8, C to Gene9).