FIG. 1.
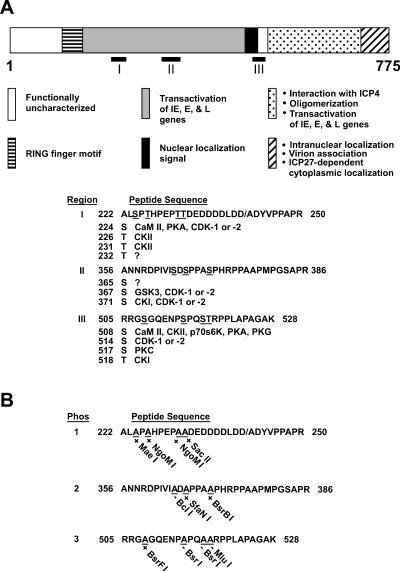
Putative phosphorylated sequences on ICP0. (A) Location of phosphorylated sequences on ICP0 as determined by μLS-MS/MS analysis. The 775 amino acids of ICP0, the locations of the major functional domains of ICP0, and the locations of the 11 putative phosphorylation sites in regions I, II, and III are shown. Beneath the diagram of ICP0, the amino acid sequences of regions I, II, and III are shown. The slash mark between D and A in region I represents the amino acid boundary between the second and third exons of ICP0. The codon numbers of the putatively phosphorylated serine (S) and threonine (T) residues are listed beneath the peptide sequence in each region. To the right of each serine or threonine are listed the cellular kinases which most likely target these residues, as determined by computer-based modeling with NetPhos 2.0 (Center for Biological Sequence Analysis, Technical University of Denmark), ScanProsite (Swiss Institute for Bioinformatics), and MacVector 7.1.1 (GCG, Madison, Wis.). Kinases include calmodulin kinase II (CaM II), protein kinase A (PKA), cyclin-dependent kinase 1 (cdk-1) and 2, casein kinase I (CKI), glycogen synthase kinase 3 (GSK3), casein kinase II (CKII), p70S6K kinase, protein kinase G (PKG), and protein kinase C (PKC). (B) Mutations in putative ICP0 phosphorylation sites. Serines and threonines in each phosphorylated region were mutated to alanine (A), resulting in the gain (+) or loss (−) of a specific restriction enzyme cleavage site, as shown beneath each substituted alanine residue. Forms of ICP0 in which phosphorylated serine (S) or threonine (T) residues in regions I, II and III have been changed to alanine (A) are designated Phos 1, 2, and 3, respectively, in the text.