Figure 1.
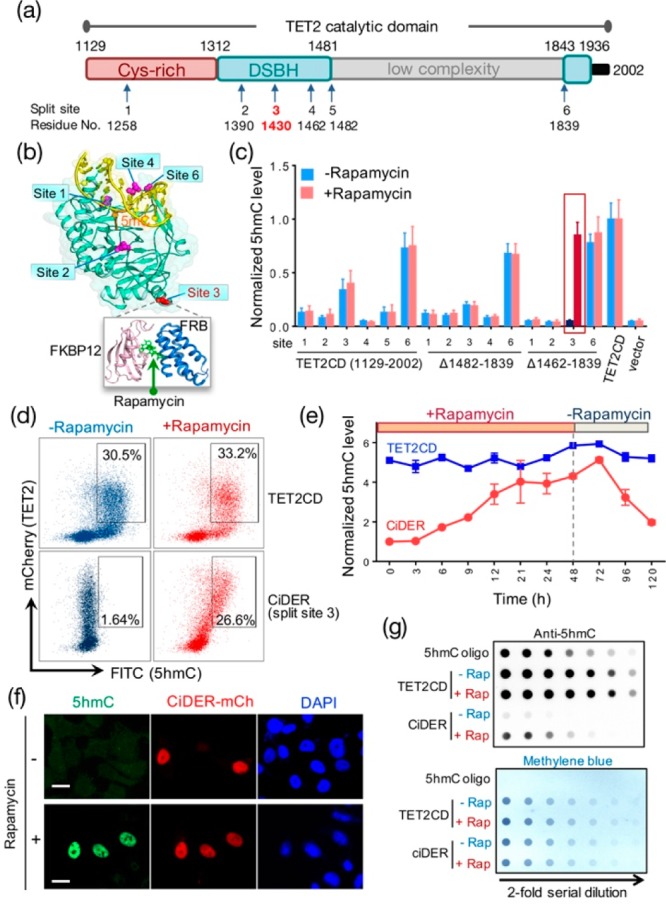
An engineered split-TET2 enzyme for inducible DNA hydroxymethylation in mammalian cells. (a) Domain architecture of the catalytic domain of TET2 (TET2CD; aa 1129–2002) and positions of selected split sites. DSBH, double stranded beta helix. (b) Split sites mapped to the 3D structure of TET2CD (PDB entry: 4NM6). A rapamycin-inducible heterodimerization module composed of FKBP12 and FRB was inserted individually into the selected split sites. (c) Screening and optimization of split-TET2CD constructs to achieve chemical-inducible 5hmC generation in HEK293T cells. The construct with insertion of FKBP12-T2A-FRB at split site 3 and deletion of the low complexity region (Δ1462–1839) stood out as the best candidate (termed “CiDER”). AP1903-incucible homodimerization of a mutant FKBP12 (F36 V) can also be engineered into this position to restore the catalytic activity of split-TET2CD (Figure S2). (d) Quantification of CiDER-mediated 5hmC production by flow cytometry. HEK293T cells transfected with mCherry (mCh)-tagged CiDER or mCh-TET2CD (positive control) were immunostained with an anti-5hmC primary antibody and an FITC-labeled secondary antibody. (e) Time course of rapamycin (200 nM)-induced production of 5hmC in HEK293T cells expressing CiDER or TET2CD (as positive control). Rapamycin was washed away 48 h after incubation with cells. (f) Representative fluorescent images of 5hmC (green), CiDER-mCh (red), and nuclear staining with DAPI (blue) in HEK293T cells before and after rapamycin (200 nM) treatment. (g) Dot-blot assay to quantify rapamycin (200 nM)-induced changes of 5hmC levels in genomic DNA purified from HEK293T cells expressing CiDER or TET2CD. A synthetic oligonucleotide with a known amount of 5hmC was used as a positive control. The loading control was shown in the bottom panel by methylene blue staining of total amounts of input DNA. See Supporting Information (Figures S1,2) for more results and sequences. n = 5. Scale bar = 10 μm.
