Figure 2.
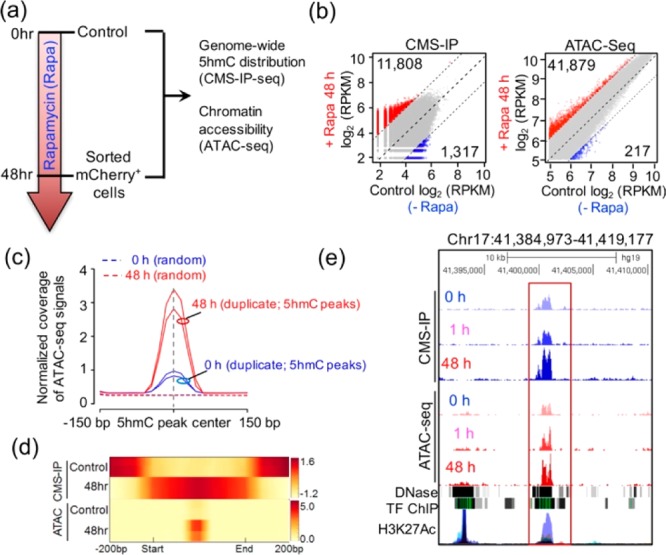
CiDER-mediated chemical inducible remodeling of epigenetic states to increase chromatin accessibility in mammalian cells. (a) Schematic of the experimental setup 0 or 48 h after 200 nM rapamycin treatment, genomic DNA samples from HEK293T cells were subjected to genome-wide 5hmC profiling and ATAC-seq to monitor chromatin accessible regions in the genome. (b) Scatter plots of differential 5hmC peaks and ATAC peaks between 0 and 48 h treatment groups. Red and blue dots represent significantly up- or down-regulated ATAC/5hmC peaks at 48 h, respectively. The gray dots represent peaks without significant changes at 48 h. (c) Normalized coverage of ATAC-seq signals (0 h and 48h; with biological duplicates) were plotted 150 bp up- and downstream of the centers of 5hmC-peaks at 0 h (control). (d) Distribution of averaged 5hmC enrichment (2 upper panels) and ATAC-seq peak enrichment (2 lower panels) at control ATAC/5hmC peaks regions in CiDER-expressing HEK293T cells at time 0 (control) or 48 h following rapamycin treatment. (e) Representative genome browser view of one HMDR in the LINC00854 locus on chromosome 17 that showed rapamycin-induced gain of 5hmC and ATAC-seq peaks, which was overlaid with traces representing DNase I hypersensitive sites, transcriptional factor binding (TF ChIP) and H3K27Ac enrichment.
