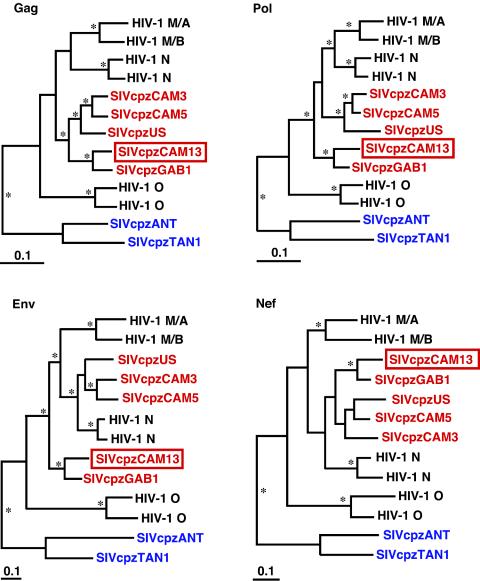
FIG. 5.
Phylogenetic trees depicting the relationship of SIVcpzCAM13 to other HIV-1 and SIVcpz isolates. SIVcpz strains from P. t. troglodytes are shown in red, while those from P. t. schweinfurthii are shown in blue; the new SIVcpzCAM13 is boxed. Deduced amino acid sequences of SIVcpzCAM13 were compared with the sequences of SIVcpzGAB1 (GenBank accession number X52154), SIVcpzUS (AF103818), SIVcpzCAM3 (AF115393), and SIVcpzCAM5 (AJ271369) and of HIV-1 group N strains YBF30 (AJ006022) and YBF106 (AJ271370), group O strains ANT70 (L20587) and MVP5180 (L20571), and group M subtypes A (strain U455; M62320) and B (LAI; K02013). Sequences were aligned by using CLUSTAL W (28), with minor manual adjustment. Trees were estimated by the Bayesian method (implemented in MRBAYES [12]), by using the JTT matrix of amino acid replacement (15) and a gamma distribution of rates across sites and then midpoint rooted. Asterisks denote branches with estimated posterior probabilities of at least 95%. The scale bars indicate 0.1 replacements per site.