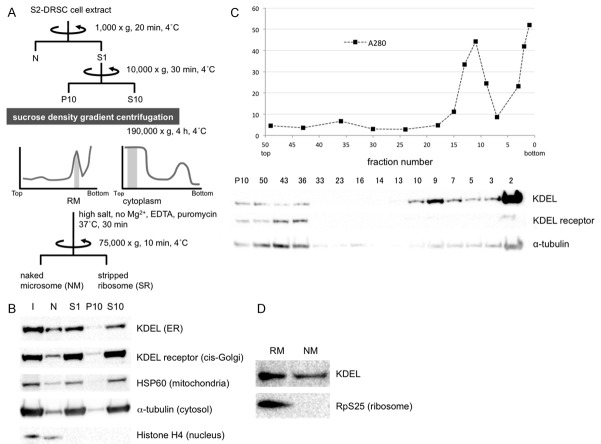
Figure 1.
Rough microsomal (RM), cytoplasm (C), naked microsome (NM) and stripped microsome (SR) fractions were purified from S2-DRSC cells to identify mRNAs targeted to ER in an SRP-independent manner. A. Scheme for isolation of RM, C, NM and SR fractions. Shaded regions in the graphs indicate fractions that gave rise to the RM and C pools. B. After fractionation, organelle distributions were determined by Western blot analysis with anti-KDEL (ER), KDEL receptor (cis-Golgi), HSP60 (mitochondria), a-tubulin (cytosol) and Histone H4 (nucleus) antibodies, respectively. I = input. C. P10 fraction by sucrose density gradient centrifugation. The graph shows total protein levels detected by UV spectrometry, and the Western blot below illustrates the separation of ER and Golgi. D. Ribosomes on ER-containing fractions were stripped by treatment with EDTA and puromycin in high salt buffer. As expected, the ribosomal marker RpS25 detected by Western blot analysis is no longer detected in the NM fraction.