Figure 3.
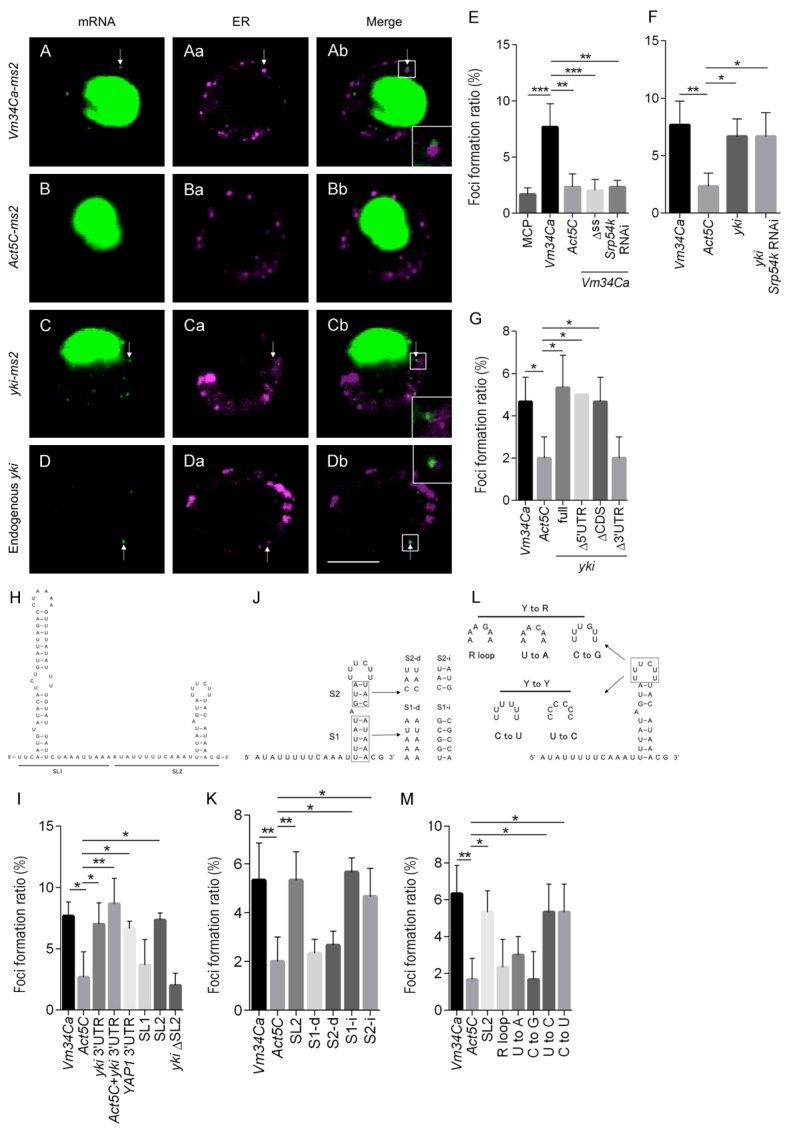
yki mRNA forms foci in S2-DRSC cells in the vicinity of the ER via the SL2 motif in the yki 3’UTR. (A-D) Subcellular RNA localization. (Aa-Da) The ER is marked by ss-DsRed-HDEL. Scale bar, 5 μm. S2-DRSC cells expressing target-ms2 RNA, NLS-MCP-GFP and ss-DsRed-HDEL (A-C) were analyzed by confocal microscopy. While Vm34Ca-ms2 (A-Ab) and yki-ms2 (C-Cb) RNA form foci (arrow), Act5C-ms2 RNA (B-Bb) rarely forms foci. ss-DsRed-HDEL marks ER. NLS = nuclear localization signal; ss = signal sequence; HDEL = ER retention signal. (D-Db) Endogenous yki mRNA and ER are detected by in situ hybridization and anti-KDEL antibody immunostaining, respectively. (Ab-Db) RNA foci are localized adjacent to the ER (white rectangle is magnified in inset, bottom right). (E-G, I, K, M) The foci formation ratio was calculated from the number of cells forming foci in the cytoplasm to the total number of cells expressing GFP in the nucleus. Data are shown as means ± SD (n = 3). (E) Vm34Ca and Act5C-ms2 RNAs were used as positive and negative controls, respectively. Vm34Ca-ms2 RNA produces foci in an SRP-dependent manner. (F, G) The yki-ms2 RNA also forms foci efficiently, but in an SRP-independent, and 3’UTR-dependent manner. (H, I) SL2 in the yki 3’UTR is necessary for foci formation. (H) Predicted secondary structure of the yki 3’UTR: SL1 = stem loop1; SL2 = stem loop2. (J-M) The structure of the SL2 stem regions and pyrimidines in the loop region are critical for foci formation. R = purine; Y = pyrimidine. (J) S1-d and S2-d mutations in SL2 are intended to disrupt secondary structure, while S1-i and S2-i mutations are not. (K) High foci formation ratio is dependent on SL2 secondary structure. (L) Bases in the loop region were exchanged from pyrimidine to purine or from pyrimidine to pyrimidine. (M) The introduction of purines in the loop region decreases foci formation. Data are shown as means ± SD (n = 3). *P<0.05, **P<0.01, ***P<0.001 (one-way ANOVA, Dunnett’s Multiple comparisons test).
