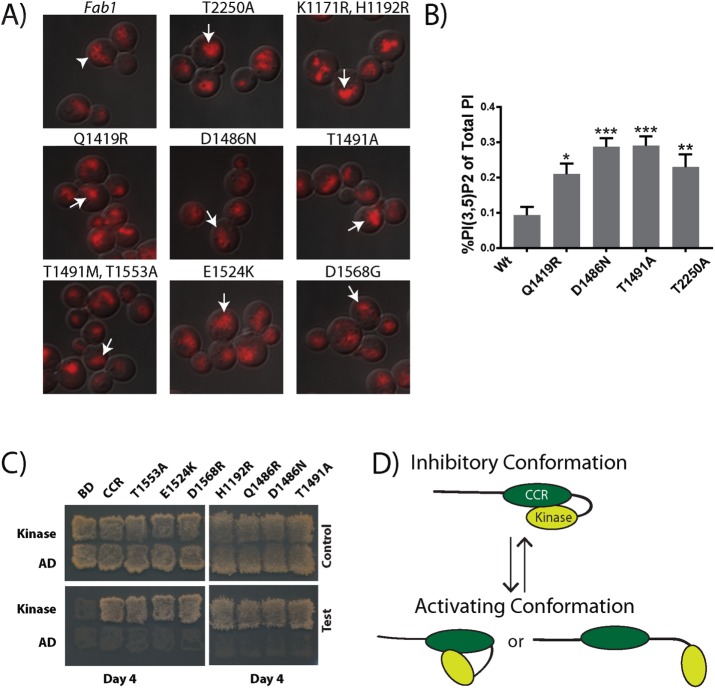
FIGURE 5:
Dominant active mutations in the CCR region of Fab1 indicate a regulatory role for this domain. (A) Selected CCR point mutants exhibit a fragmented vacuole phenotype (arrows) relative to the wild-type Fab1 control (arrowhead), suggesting an elevation in PI(3,5)P2 levels. Vacuoles labeled with FM4-64 (red). Three independent experiments. (B) Under basal conditions, each of the three CCR domain mutants tested (Fab1Q1419R, Fab1D1486N, and Fab1T1491A) displays elevated levels in PI(3,5)P2 relative to Fab1WT, similar to the kinase domain mutation Fab1T2250A. Statistical significance of observed differences between wild-type and the mutant alleles was analyzed by one-way ANOVA, p < 0.0001, with a Dunnett's multiple-comparison post hoc test: *p < 0.01, **p < 0.001, ***p < 0.0001. Error bars represent SD. Three independent experiments. (C) Dominant-active mutations identified in the CCR region do not block the Y2H interaction of the CCR region with the kinase region. Y2H tests of Gal4-AD fused with the kinase region (residues 1723–2278) with Gal4-BD fused with either the CCR region (residues 1181–1585) or this region with one of the mutations H1192R, Q1419R, D1486N, T1491A, T1491M, E1524K, T1553A, or D1568R. Six independent experiments. (D) Proposed regulatory intramolecular interaction within Fab1. The CCR domain interacts with the kinase region of Fab1 and reversibly inhibits its activity. Potential CCR inhibition of Fab1 kinase activity may be relieved by posttranslational modifications and/or altered interactions with other proteins.