Figure 4.
In Vivo TRM4B-Dependent Substrate Recognition.
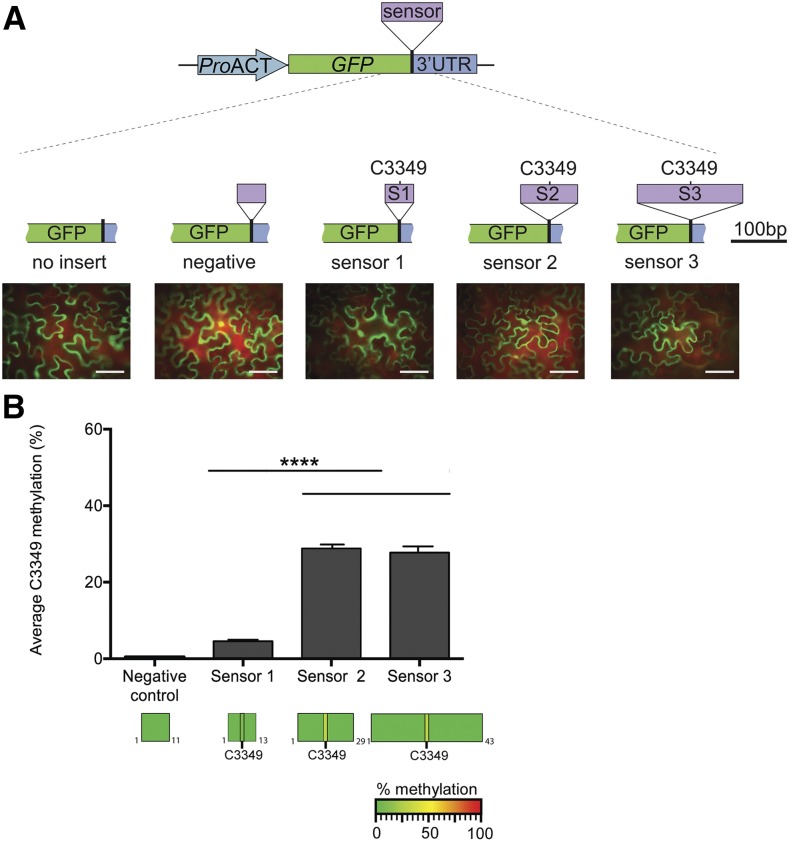
(A) Schematic representation of m5C methylation sensor constructs. Three different sized sequences flanking the MAG5 C3349 methylated site were cloned in between the GFP coding region and the beginning of the 3′UTR sequence and are referred to as sensor 1 (S1; 51 nucleotides), sensor 2 (S2; 101 nucleotides), and sensor 3 (S3; 189 nucleotides). The C3349 methylated site is located in the center of all three sensor fragments. A fourth region, derived from MAG5 exon one, was used as the “negative” control in addition to a “no insert” empty vector control. Representative images of N. benthamiana leaves transiently expressing GFP reporter for each of the sensor constructs are shown. Bar = 100 µm.
(B) Cytosine methylation in sensor fragments identified by bsRNA-amp-seq. Top: Average MAG5 C3349 methylation percentage in sensors 1 to 3 from three biological replicates. Each biological replicate was constructed by purifying RNA from three infiltrated leaves of N. benthamiana, and the RNA was then bisulfite treated, cDNA synthesis performed, and sensor regions were PCR amplified and Illumina sequenced. No nonconverted cytosines were identified in the negative control construct. Error bars indicate ± se of the mean (****P < 0.001, one-way ANOVA; n = 3). Bottom: Heat maps showing the cytosine methylation status within sensor fragments. No nonconverted cytosines were identified in the flanking sequences.