Figure 7.
Oxidative Stress-Responsive Pathways Are Constitutively Activated in trm4b Mutants.
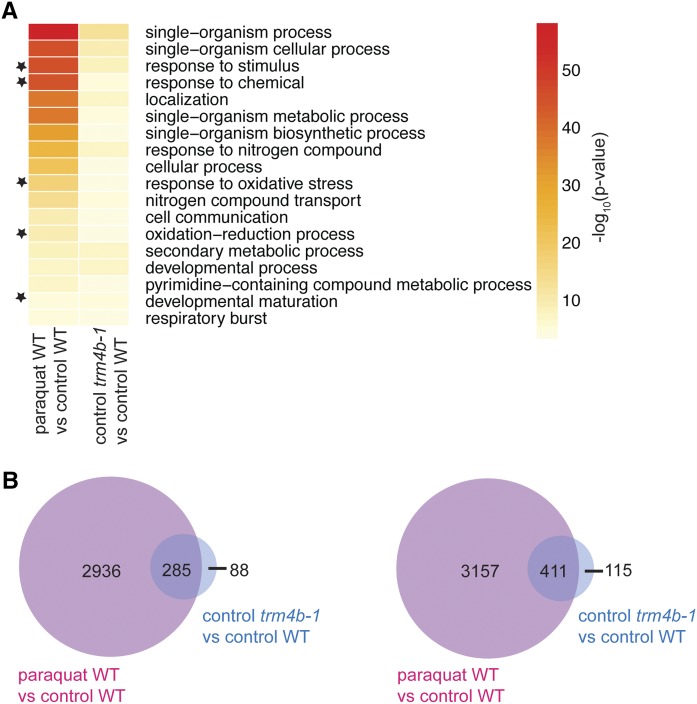
Whole roots at 6 DAG were harvested from wild-type, trm4b-1, and paraquat treated wild-type seedlings, and RNA-seq analysis was performed (n = 3). Most genes differentially expressed in trm4b-1 mutants compared with the wild type are also differentially expressed in paraquat-treated wild-type plants compared with control conditions.
(A) Biological process GO terms involved in the oxidative stress response are significantly enriched (FDR < 0.05) in trm4b-1 mutants compared with the wild type and in paraquat-treated wild-type plants compared with untreated wild-type plants. GO terms significantly enriched in both comparisons are depicted, with black stars indicating GO terms relating to oxidative stress responses. Heat map shows the significance level using the negative log of the P value, where red = very significant and yellow = significant (GO term FDR < 0.05).
(B) Venn diagrams show the overlap of differentially upregulated (left) and downregulated (right) genes (FDR < 0.05).