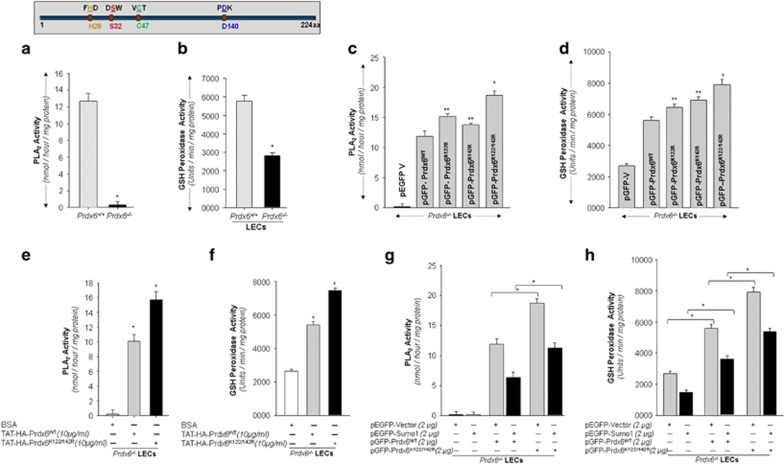
Figure 7.
(a and b). Prdx6-deficient LECs displayed insignificantly low levels of Phospholipase A2 as well as lower GSH peroxidase activities compared with Prdx6+/+. Prdx6+/+ and Prdx6−/− LECs cultured in identical conditions as described in 'Materials and methods'. Cells were harvested and total extracts containing equal amounts of proteins were processed to measure PLA2 (a) and glutathione peroxidase activity (b) following the company's protocols. Black bars show significantly reduced PLA2 and GSH peroxidase activities in Prdx6-deficient cells. The data represent the mean±SD from three independent experiments (*P<0.001). Upper panel, a schematic illustration of active sites responsible for PLA2 (S32/H26/D140) and GSH peroxidase (C47) activities. (c and d). Disruption of Sumoylation motif K122/142 R in Prdx6 protein promoted PLA2 and glutathione peroxidase activities. Prdx6−/−LECs were transfected with pEGFP-Vector, pGFP-Prdx6WT and its mutants, pGFP-Prdx6K122R, pGFP-Prdx6K142 R and pGFP-Prdx6K122/142R fused to GFP plasmids. After 48 h, total lysates containing equal amounts of proteins were processed for PLA2 (c) and GSH peroxidase (d) activities through Enzchek PLA2 and GSH peroxidase assay kits (Invitrogen), respectively. (Prdx6WT versus mutants; *P<0.001; **P<0.05). (e and f). Recombinant mutant Prdx6K122/142 R protein had increased PLA2 and GSH peroxidase activities compared with Prdx6WT. Prdx6−/−LECs were transduced with TAT-HA-Prdx6 and its mutant TAT-HA-Prdx6K122/142 R. After 24 h, total protein was isolated and assays were performed for PLA2 (e) and Glutathione peroxidase (f) activities as described in ‘Materials and methods' section. The data represent the mean±SD from three independent experiments (*P<0.001). (g and h). Sumo1 overexpression diminished phospholipase PLA2 and GSH peroxidase activities. Prdx6−/−LECs were co-expressed with pEGFP-Sumo1 along with pEGFP-Vector or pGFP-Prdx6WT or its mutant K122/142 R, 48 h later total cell lysates containing equal amounts of proteins were utilized to measure PLA2 (g) and glutathione peroxidase (h) activities; results are presented as nmol/min/mg protein and units/min/mg protein, respectively. The data represent the mean±SD from three independent experiments (*P<0.001)