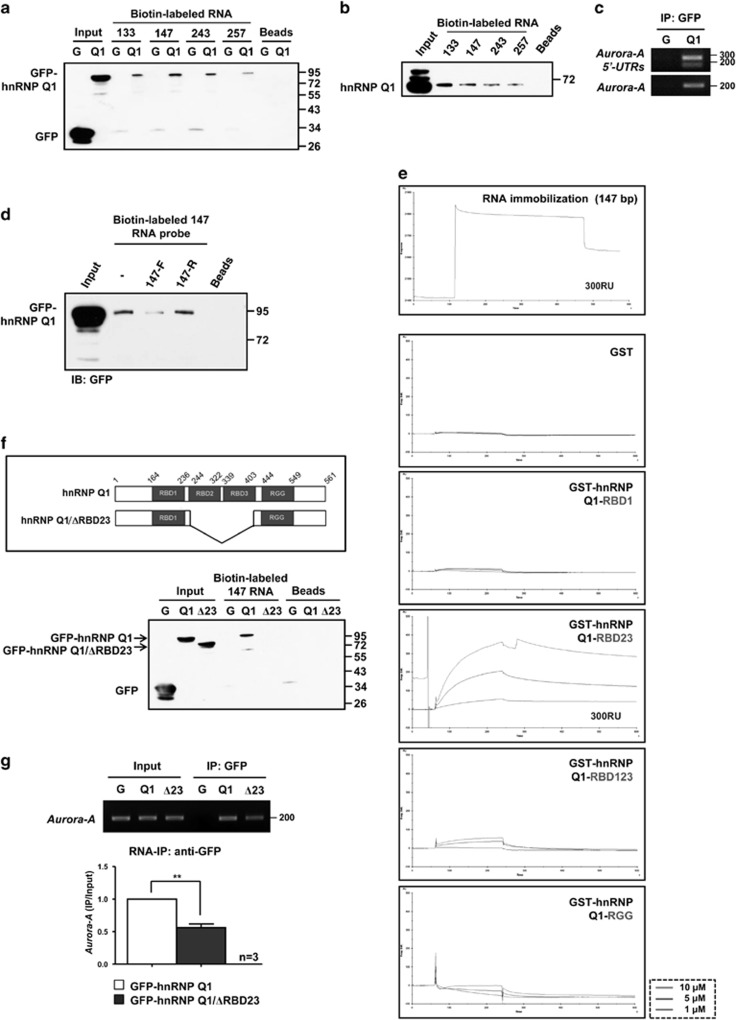
Figure 2.
HnRNP Q1 directly binds to Aurora-A mRNA 5′-UTR through RBD23 domains. (a and b) Four types of biotin-labeled Aurora-A mRNA 5′-UTR RNA probes – 133 nt, 147 nt, 243 nt or 257 nt were incubated with total cell lysates from GFP (G) or GFP-hnRNP Q1 (Q1)-expressing cells (a) or SW480 cells (b). The pulled-down proteins were separated by SDS-PAGE and immunoblotted with anti-GFP or anti-hnRNP Q antibody. (c) Cytoplasmic lysate from GFP (G) or GFP-hnRNP Q1 (Q1)-expressing cells were collected for the RNA-immunoprecipitation (RNA-IP) assay using anti-GFP antibody. The precipitants were collected for RNA extraction and subjected to RT-PCR to assess the binding of Aurora-A mRNA or Aurora-A 5′-UTRs. (d) Biotin-labeled Aurora-A mRNA 5′-UTR 147 nt RNA probe was incubated with cell lysates from GFP-hnRNP Q1-expressing cells in the presence of a twofold amount of unlabeled Aurora-A mRNA 5′-UTR 147 nt forward RNA probe (147-F) or reversed 147 nt RNA probe (147-R) as described in a. (e) Biotin-labeled Aurora-A mRNA 5′-UTR RNA probe (147 nt) was immobilized on a streptavidin (SA) sensor chip, and then GST-hnRNP Q1 proteins were subjected to an in vitro binding assay by SPR. The interaction between the Aurora-A 5′-UTR and GST-hnRNP Q1 proteins was quantified by the change of resonance units (RU) on the chip. Three different concentrations of GST-hnRNP Q1 proteins, 1 μM, 5 μM and 10 μM, were used. (f) (Upper) Scheme illustrates RBD23-truncated hnRNP Q1 (GFP-hnRNP Q1/ΔRBD23). (Lower) Total cell lysates from GFP (G), GFP-hnRNP Q1 (Q1) or GFP-hnRNP Q1/ΔRBD23 (Δ23)-expressing cells were collected for biotin pull-down assay as described in a. (g) RNA-IP assay to investigate the interaction between GFP (G), GFP-hnRNP Q1 (Q1) and GFP-hnRNP Q1/ΔRBD23 (Δ23) with Aurora-A mRNA was performed as described in c. The associated Aurora-A mRNA with hnRNP Q1 was detected by PCR (upper) and real-time qPCR (lower); **P<0.01