Figure 1. Identification of yeast genes mediating response to UV, X-ray radiation, or cisplatin.
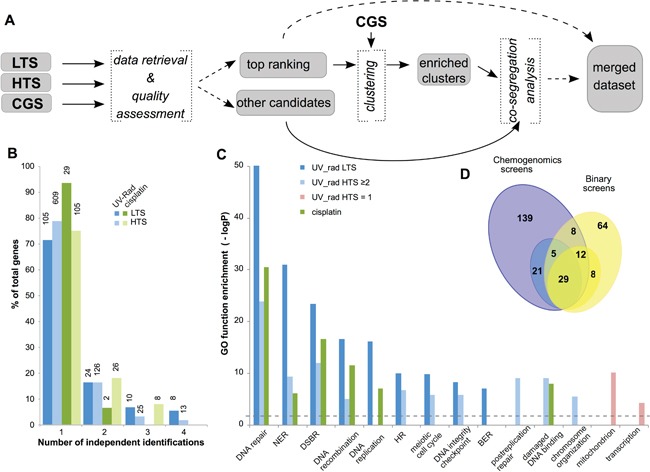
A. Workflow for identification of yeast genes modulating sensitivity to UV, ionizing radiation, and cisplatin treatment. LTS, low throughput screen; HTS, high throughput screen; CGS, chemogenomic screen. B. Classification of candidate genes based on the number of independent studies that identified each gene as contributory to resistance to UV or radiation (UV_rad) or to cisplatin, as well as by type of supporting study (LTS and HTS). Y-axis indicates percent of genes for each class, while the absolute number of supporting studies is shown on the top of each bar. C. Enrichment in GO terms for specific subsets of candidate genes. NER, nucleotide excision repair; DSBR, double strand break repair; HR, homologous recombination; BER, base excision repair. Dashed line indicates the threshold for the statistical significance. D. Overlap between cisplatin resistance gene sets identified in binary LTS/HTS (yellow) and chemogenomic (blue) screens: numbers represent individual genes. Darker colors indicate genes supported by ≥2 binary screens, or by 3 independent chemogenomic studies; lighter colors indicate genes supported by 1 binary screen, or by 2 of 3 chemogenomic studies.
