Figure 2. Examples of functional clusters of genes regulating response to UV, X-ray radiation, or cisplatin.
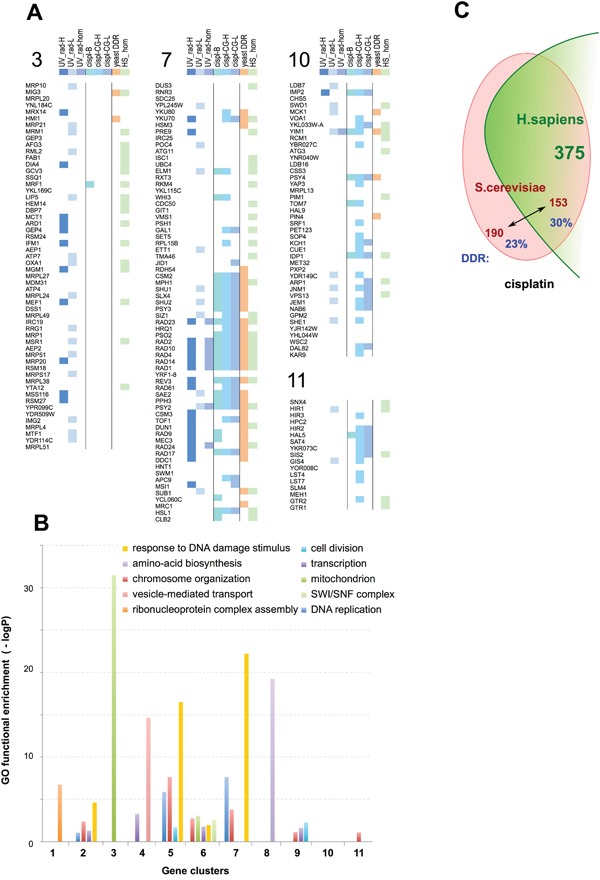
A. Graphical representation of composition and selected properties of clusters (CL) enriched in UV_rad sensitivity-modulating genes (CL 3), in cisplatin sensitivity-modulating genes (CL 10 and 11), or in both (CL 7). Key indicates genes identified as UV_rad HTS≥2 and/or UV_rad LTS (UV_rad-H); or as inducing cisplatin resistance from binary (cispl-B), or chemogenomics (cispl CG-H) with high statistical significance (see text for details). Genes initially defined as lower confidence because of identification from the UV_rad HTS=1 set alone, or only by a single high score for cisplatin sensitization from a single chemogenomics screen, but significantly enriched (p<0.05) within clusters, are denoted as “UV_rad-L” and “cispl-CG-L”, respectively. “Yeast DDR” indicates genes annotated as involved in response to DNA damage; “UV_rad-hom” indicates functional orthologs in fly and/or worm; “HS_hom”, genes have unambiguous orthologs in H. sapiens. See Supplementary Table 3 for detailed information on all identified clusters. B. Enrichment in Gene Ontology (GO) functions for the identified clusters. C. Overall evolutionary conservation of S. cerevisiae cisplatin resistance genes in H. sapiens. Numbers shown in red font represent individual yeast genes, while green font is used for the number of human genes orthologous to the yeast counterpart. The fraction of yeast genes annotated as involved in DDR is shown in blue.
