Figure 2. miR-320a functions as a glioma suppressor by directly targeting SND1 and β-catenin.
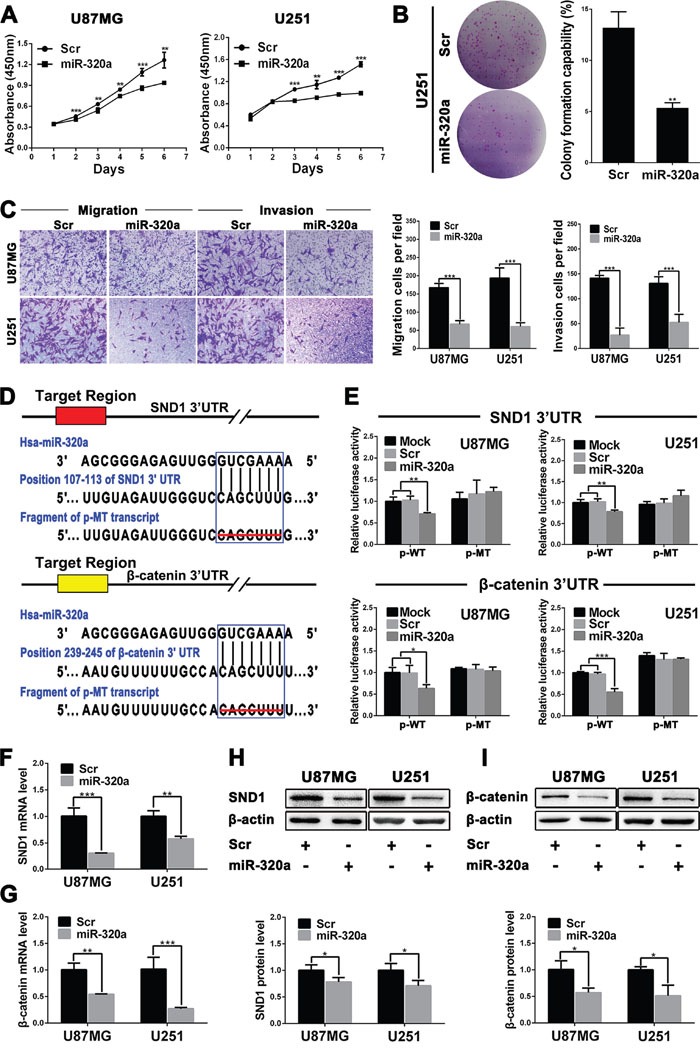
A. Growth curves from U87MG and U251 cells transfected with scrambled control sequence (Scr) or miR-320a mimics (miR-320a) assessed by CCK8 assay. B. Colony formation efficiencies of the transfected U251 cells. C. Representative images (left) and numbers (right) of the indicated migratory and invasive cells analyzed by transwell assay. D. Predicted miR-320a binding sites in the 3′-UTRs of SND1 and β-catenin by TargetScan and the designed mutant 3′-UTRs in which miR-320a binding sites were deleted. E. Dual luciferase reporter assays in U87MG and U251 cells transfected with p-WT or p-MT alone (Mock) and cotransfected with p-WT or p-MT and Scr or miR-320a; p-WT=the reporter plasmid of wild-type SND1 or β-catenin 3′-UTR, p-MT=the reporter plasmid of mutant-type SND1 or β-catenin 3′-UTR. The data were normalized according to the ratio of firefly luciferase activity to renilla luciferase activity. F and G. SND1 and β-catenin mRNA levels in the indicated cells analyzed by qRT-PCR and normalized against GAPDH. The ratios of SND1/GAPDH and β-catenin/GAPDH in the Scr-transfected cells were arbitrarily set to 1.0. H and I. SND1 and β-catenin protein levels in the indicated cells detected by Western blot and normalized against β-actin. All the experiments were performed at least in triplicate and the data are presented as the mean ± SD. * P<0.05, ** P<0.01, *** P<0.001.
