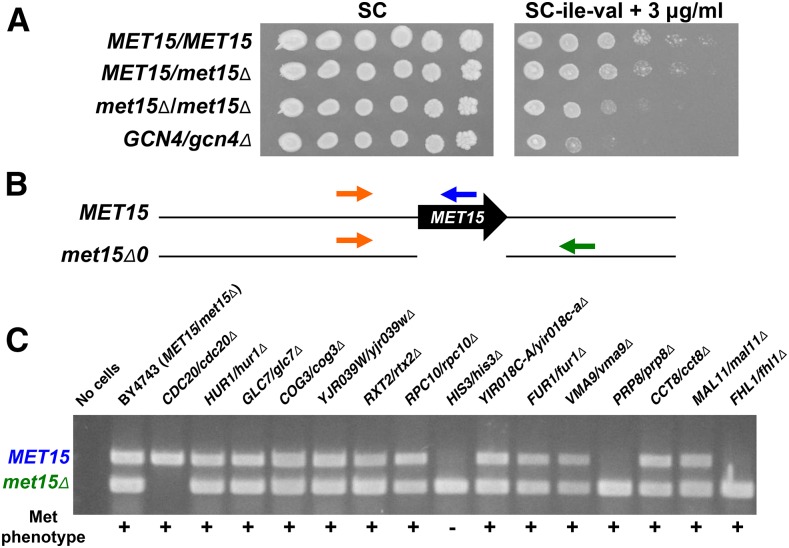
Figure 3.
Loss of heterozygosity at the MET15 locus and effects on SMM phenotypes. (A) Homozygous met15∆ strains are sensitive to SMM. The MET15/MET15, MET15/met15∆ (met15Δ0; the strain is BY4743), met15∆/met15∆ (met15Δ::kanMX4/met15Δ0, which is the met15Δ strain from the heterozygous collection), and GCN4/gcn4∆ strains were grown in YPD in a 96-well microtiter for 2 d at 30°. Ten-fold serial dilutions were made and 5 µl of undiluted and diluted samples were spotted onto SC control and SC-ile-val + 3 µg/ml SMM media. The plates were incubated at 30° and photographed after 3 d of growth. (B) A diagram of the MET15 locus and oligonucleotides for yeast colony PCR are shown. The primer depicted by the orange arrow (upstream primer, see Materials and Methods) is upstream of the MET15 locus and will bind to both MET15 and met15∆ alleles. The primer indicated by the blue arrow (ORF primer, see Materials and Methods) binds to the MET15 coding region, and it will not bind to the met15∆ allele. With the upstream primer, the MET15 allele will yield a PCR product of ∼1 kbp. The primer depicted by the green arrow (downstream primer, see Materials and Methods) binds to a region beyond the MET15 stop codon. This region is present in both the MET15 and met15∆ alleles, but conditions for PCR were performed such that only the shorter, met15∆-generated PCR product was amplified. (C) A representative gel of the MET15 locus PCR from candidate and control strains is shown. Sample names are listed above the agarose gel image. Negative (no cells) and positive (BY4743 with both the MET15 and met15∆ alleles) controls were included in each gel. Methionine phenotypes are listed for each strain below the agarose gel (+, methionine prototroph; −, methionine auxotroph). Several strains show LOH at the MET15 locus as indicated by a single band on the gel.