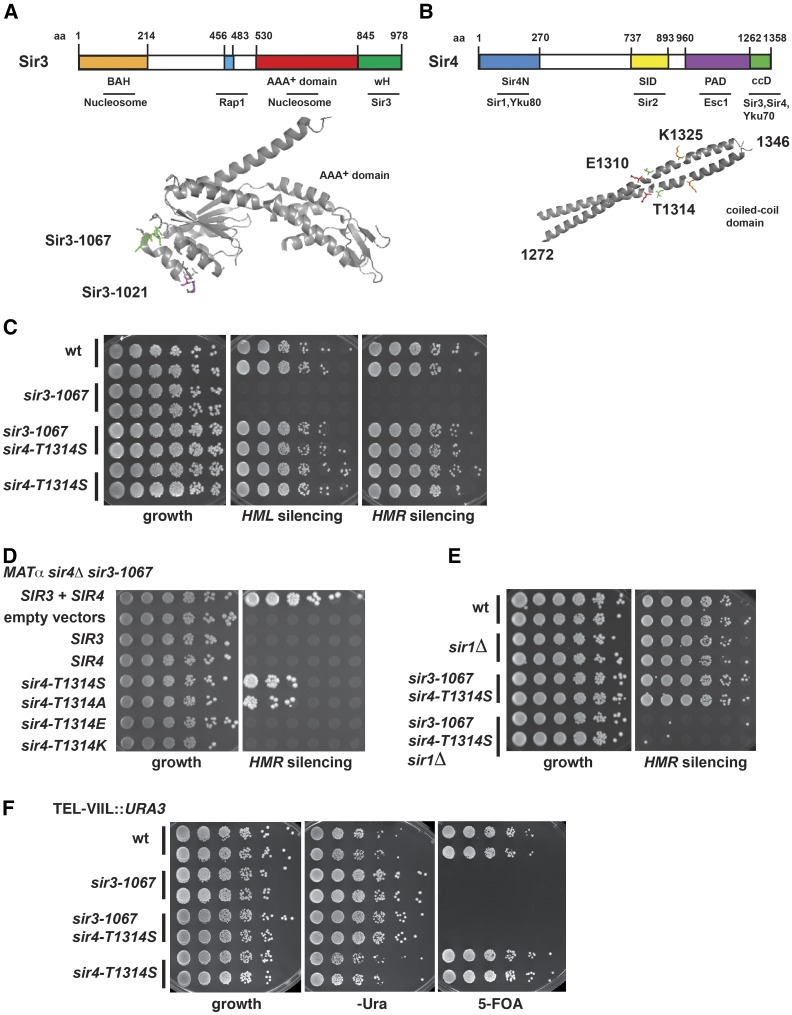
Figure 1.
Identification of a mutation in the coiled-coil domain of Sir4 that suppresses the HM silencing defect of a mutation in the Sir3 AAA+ loop. (A) Schematic illustration of the Sir3 protein (top), and structure of the AAA+ ATPase-like domain (below). BAH, bromo-adjacent homology domain; wH, winged-helix domain. The sir3 alleles sir3-1067 (K657A, K658A) and sir3-1021 (D640A, S642L) are mapped on the structure (PDB: 3TE6) (B) Schematic illustration of the Sir4 protein including the C-terminal coiled-coil domain (ccD, aa 1262–1358), the Sir2 interaction domain (SID), and the partitioning and anchoring domain (PAD) (top). Below, structure of the Sir4 coiled-coil domain. The amino acids E1310, T1314, and K1325 that are relevant for this study are mapped on the structure (PDB: 1PL5). (C) Mutation of Sir4-T1314 to serine (sir4-T1314S) restored the silencing defect of sir3-1067 at HMR (AEY5554) and HML (AEY5555). A semiquantitative mating assay was performed as described in Materials and Methods, and plates were incubated for 3 d at 30°. (D) Mutation of Sir4-T1314 to serine and alanine (sir4-T1314S, sir4-T1314A), but not glutamine (E) or lysine (K), restored the HMR-silencing defect of sir3-1067. Plasmids encoding the respective Sir3 and Sir4 versions were transformed into a MATα sir3-1067 sir4Δ strain (AEY5184). (E) Suppression of sir3-1067 by sir4-T1314S depended on Sir1. (F) Sir4-T1314S did not suppress the telomeric silencing defect of sir3-1067. sir3-1067 and sir4-T1314S were chromosomally integrated into a TEL-VIIL::URA3 strain, and serial dilutions were spotted on a 5-FOA containing plate to analyze their ability to silence the URA3 reporter gene at the telomere. Cells were additionally spotted on supplemented minimal medium as a growth control. The plates were incubated for 3 d at 30°.