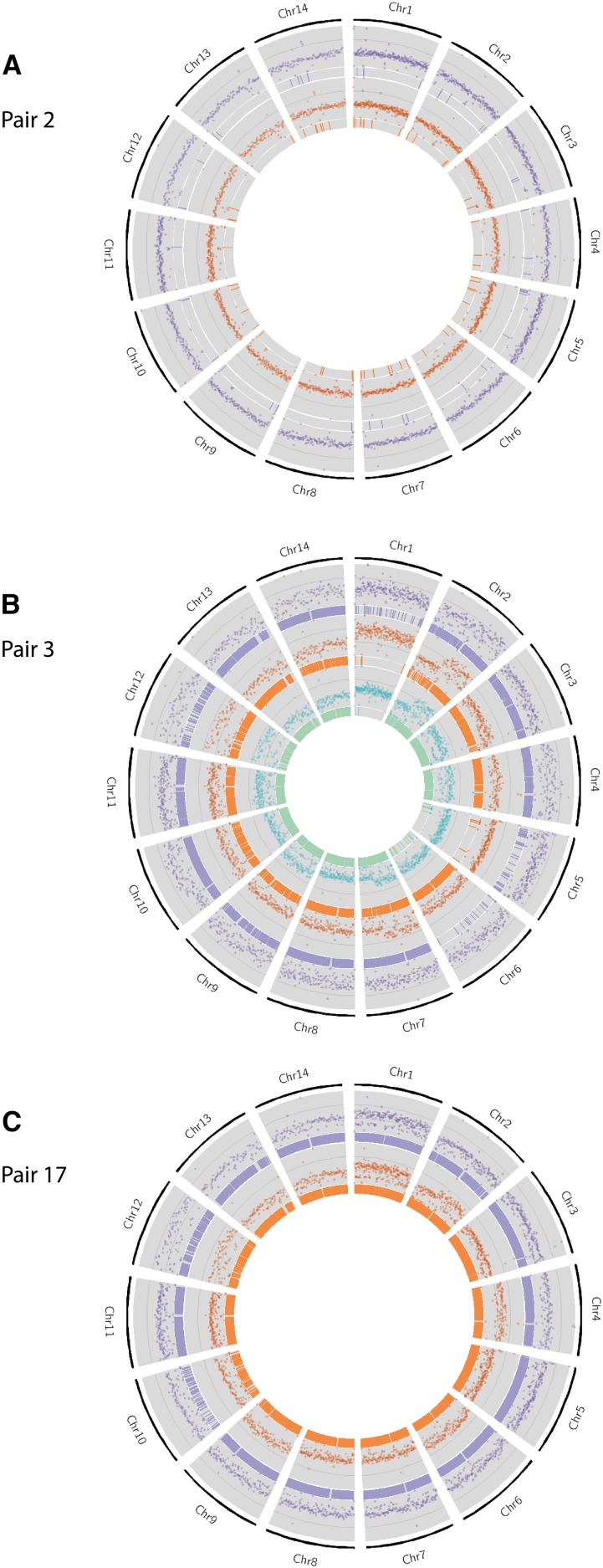
Figure 2.
Extensive chromosomal copy number variation was observed in all isolates in pairs 3 and 17, when compared to H99. Pair 2 is included to illustrate isolates without ploidy and extensive nsSNPs. Here, normalized whole-genome depth of coverage is shown, averaged over 10,000 bp bins, in scatter plots. Bar plots represent the position of nsSNPs. The purple track represents the original isolate, orange the recurrent isolate, and green (in the case of pair 3) for the final recurrent isolate. (A) No increase in ploidy is observed in either the original or recurrent isolate of Pair 2, and a small number of nsSNPs are seen. (B) Increase-in-ploidy is observed in many chromosomes in the day 1 isolate for pair 3, some of which are lost over time. A large number of nsSNPs are observed in all chromosomes in isolates of pair 3, with Chr 6 being the exception: very few nsSNPs are located in Chr 6 in CCTP50 and CCTP50-d409, whereas over 2000 nsSNPs are observed in Chr 6 in CCTP50-d257. (C) A gain in ploidy is observed for Chrs 2, 4, 6, and 9 compared to the day 1 isolate in pair 17, whereas ploidy remains unchanged for Chr 1 and 12. Chr, chromosome; nsSNP, nonsynonymous single nucleotide polymorphism.