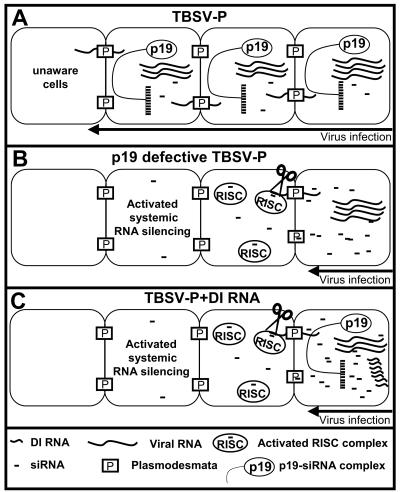
FIG. 5.
Proposed model for the PTGS-based mechanism of DI RNA-mediated interference. (A) In wild-type virus infections, the viral genomic RNA accumulates to high levels in infected mesophyll cells and the PTGS machinery of the host produces virus-specific siRNAs. However, at the same time, the virus translates p19, which physically binds the generated siRNAs, inhibiting their spread out of the infected cell. Consequently the cells ahead the infection front remained unprotected against the spreading virus. (B) Cells infected with p19-defective virus accommodate the same level of virus genomic RNA as cells infected with wild-type virus because RNA silencing has no capacity to cope with the invasive accumulation of virus-derived products. However, the absence of p19 results in the accumulation of free virus-specific siRNAs, which are able to traffic through plasmodesmata to cells beyond the infection front. These virus-specific mobile siRNAs are incorporated into the RISC of cells ahead of the infection front. Thus the already-activated RISCs destroy the entering viral RNA before the establishment of virus replication. (C) In the presence of DI RNAs, the infected cells accumulate significant amounts of genomic RNA and p19. However, because DI RNAs are poor targets of PTGS, they accumulate to extremely high levels and provide a source for the generation of extra amounts of virus-specific siRNAs, saturating the p19-binding capacity. The remaining free siRNAs are able to induce a similar process, which has been described for p19-defective-virus infection. Other previously described DI RNA-associated factors, such as suppressed rate of accumulation of genomic RNA and reduced p19 accumulation in early stages of virus infection, may further enhance the efficiency of PTGS-based defense by increasing the amount of free siRNA. The synergistic mode of action of different factors results in restricted spread of the virus and development of attenuated symptoms.