Abstract
Nodamura virus (NoV) and Flock House virus (FHV) are members of the family Nodaviridae. The nodavirus genome is composed of two positive-sense RNA segments: RNA1 encodes the viral RNA-dependent RNA polymerase and RNA2 encodes the capsid protein precursor. A small subgenomic RNA3, which encodes nonstructural proteins B1 and B2, is transcribed from RNA1 during RNA replication. Previously, FHV was shown to replicate both of its genomic RNAs and to transcribe RNA3 in transiently transfected yeast cells. FHV RNAs and their derivatives could also be expressed from plasmids containing RNA polymerase II promoters. Here we show that all of these features can be recapitulated for NoV, the only nodavirus that productively infects mammals. Inducible plasmid-based systems were used to characterize the RNA replication requirements for NoV RNA1 and RNA2 in Saccharomyces cerevisiae. Induced NoV RNA1 replication was robust. Three previously described NoV RNA1 mutants behaved in yeast as they had in mammalian cells. Yeast colonies were selected from cells expressing NoV RNA1, and RNA2 replicons that encoded yeast nutritional markers, from plasmids. Unexpectedly, these NoV RNA replication-dependent yeast colonies were recovered at frequencies 104-fold lower than in the analogous FHV system. Molecular analysis revealed that some of the NoV RNA replication-dependent colonies contained mutations in the NoV B2 open reading frame in the replicating viral RNA. In addition, we found that NoV RNA1 could support limited replication of a deletion derivative of the heterologous FHV RNA2 that expressed the yeast HIS3 selectable marker, resulting in formation of HIS+ colonies.
The nodaviruses are among the simplest animal-infectious positive-sense RNA viruses (5). The alpha- and betanodaviruses infect primarily insects and fish, respectively. Alphanodavirus species include Nodamura virus (NoV), the type species, and Flock House virus (FHV), the first member for which infectious cDNA clones were obtained (7). The nodavirus genome is bipartite, with a total length of approximately 4.5 kb. RNA1 encodes the viral components required for RNA replication. The NoV and FHV RNA-dependent RNA polymerases (RdRps) share 44% amino acid sequence identity (18). RNA2 encodes the capsid protein precursor, which is 50% identical for NoV and FHV (18). Both genomic RNAs are packaged in the same virion. Infectious reassortant viruses, composed of RNA1 and RNA2 molecules from different nodaviral species, have been recovered for several alphanodavirus combinations, specifically all possible combinations of FHV, Black beetle virus, and Boolarra virus RNAs. However, infectious reassortant viruses between NoV and FHV were not recovered (10).
In addition to the two genomic RNAs, subgenomic RNA3 (typically 380 to 480 nucleotides [nt]) is produced from the 3′ end of RNA1 during RNA replication. FHV RNA3 encodes a small B2 protein from an open reading frame (ORF) that largely overlaps the RdRp ORF; NoV encodes two forms of B2 that differ only at their amino termini (16). The FHV and NoV B2 proteins can suppress RNA interference (RNAi), a highly conserved form of homology-dependent gene silencing triggered by double-stranded RNA, by an unknown mechanism (21, 22). Most alphanodavirus RNA3s also encode protein B1, of unknown function, in the same reading frame as the RdRp.
Nodavirus RNA replication has been studied in a variety of eukaryotes. Among the alphanodaviruses, only NoV has been shown to infect and kill mammals, specifically suckling mice and suckling hamsters (12, 33). However, upon experimental transduction, the RNAs of both NoV and FHV will replicate to very high levels in a variety of cell types, including those from mammals, insects, and higher plants (4, 30, 34). FHV remains the only animal-infectious virus that has been reported to replicate its entire genome and produce infectious virus in the yeast Saccharomyces cerevisiae (30), although several other viruses can undergo partial replication in yeast cells (2, 15, 26, 31, 35). The ability to study virus replication in a unicellular organism that can grow under strictly defined conditions greatly facilitates the investigation and utilization of nodaviruses.
Here we report that NoV also replicated to very high levels in yeast following experimental transfection of purified genomic RNA or following expression of genomic RNAs synthesized in vivo from infectious cDNAs. NoV RNA1 replicated to levels very similar to those of FHV RNA1, and NoV RNA1 mutants behaved in yeast as expected from previous work in other experimental systems (16, 17). However, wild-type (WT) NoV RNA2 accumulated in yeast to lower levels than did FHV RNA2. Mutations in the NoV RNA2 capsid ORF greatly enhanced levels of RNA2 replication. In order to test the ability of NoV to express heterologous proteins in the yeast system, genes encoding the yeast HIS3 or mammalian codon-optimized green fluorescent protein (GFP) ORFs were inserted into deletion derivatives of NoV RNA2. NoV RNA replication-dependent yeast colonies could be selected following the expression in vivo of the NoV RNA2-HIS3 replicon together with RNA1. However, these colonies arose at about 104-fold lower frequency than in the comparable FHV system. Sequence analysis of reverse transcription (RT)-PCR-amplified NoV RNA1 from one such colony revealed an arginine-to-glutamine mutation, R59Q, in the NoV B2 ORF. When NoV RNA1 encoding the R59Q mutation was used to support the NoV RNA2-HIS3 replicon, the frequency of colony formation approximated that observed in the FHV system. Finally, we tested the relative abilities of the NoV and FHV RdRps to support replication of NoV and FHV RNA2-derived replicons. The NoV RdRp supported limited replication of FHV RNA2 derivatives, but the FHV RdRp did not support replication of NoV RNA2 species. These results illustrate the utility of the yeast system as an investigational approach to several aspects of nodavirus replication.
MATERIALS AND METHODS
Cells, transformation, growth, and induction.
A Frozen-EZ yeast transformation II kit (Zymo Research, Orange, Calif.) was used to introduce plasmid DNAs into the synthetic deletion yeast S. cerevisiae strain BY4733 (MATa his3Δ200 leu2Δ0 met15Δ0 trp1Δ63 ura3Δ0) (6). Plasmids encoding appropriate selectable markers were cotransformed with experimental plasmids to allow growth of all yeast strains in media selective for LEU2 and TRP1. Yeast cells were grown at 30°C with glucose or galactose as the carbon source, as indicated. Figures and legends have been simplified to specify only the presence or absence of histidine, but all synthetic media also lacked leucine and tryptophan. To induce the GAL1 promoter, cells were grown to an optical density at 600 nm of ∼1.0 in liquid dextrose medium, pelleted, and resuspended in liquid galactose medium containing histidine and incubated for 2 days. For selection, yeast cells were induced for 2 days, and then 5 μl was used to seed 2-ml cultures of dextrose-containing liquid medium lacking histidine, and 2 days later, 5 μl was spotted onto dextrose-containing solid medium lacking histidine. Plates were incubated at 30°C for 4 days and photographed. The image colors were digitally reversed to enhance contrast.
Plasmid constructions.
Whenever PCR was used during plasmid construction, PCR-generated sequences were verified. The backbone of pN1 is YEplac112, a TRP1-marked shuttle vector (13), and is similar to that of pF1 (29). The backbone of pN2 is YEp351, a LEU2-marked shuttle vector (14), and is similar to that of the FHV-derived pF2U (28). In both plasmids, NoV cDNAs lie between the inducible RNA polymerase II (polII) GAL1 promoter and hepatitis delta virus (HDV) ribozyme cDNA, and the sequence 5′…AAAAGTACTGCAXGGGTCGG…3′, where X represents the full-length NoV cDNA, describes the promoter-cDNA-ribozyme junction. Three previously described mutant versions of the NoV RNA1 cDNA (NoV1) were introduced into pN1: U1274C contains a single point mutation at NoV RNA1 nt 1274 that decreases synthesis of RNA3 (16). B2−− contains three point mutations (U2745C, U2754C, and C2757C) and eliminates synthesis of both forms of protein B2 (16). RNA3− contains four point mutations (C2731A, G2732A, U2733A, and G2734A) that together eliminate synthesis of RNA3 (17). A frameshift mutation in the RdRp coding region had been previously introduced into the NoV RNA1 cDNA by destruction of the EcoRI site at position 982 by Klenow fill-in and religation (K. L. Johnson and L. A. Ball, unpublished data); this frameshift mutation was also introduced into pN1.
Plasmid pN2 also differs from pF2U in that it has sequences corresponding to the ADH1 polyadenylation site just 3′ of the HDV ribozyme. The Klenow-filled pAAH5 (1) HindIII-SphI fragment containing the ADH1 polyadenylation site was cloned into the Klenow-filled XbaI-SphI site of the YEp351 multiple cloning sequence, the HindIII site of which had also been destroyed by Klenow fill-in and religation. Two mutant forms of the NoV RNA2 cDNA (NoV2) were also introduced into pN2. Nucleotides 53 to 476 were deleted from pN2 and replaced by a six-nucleotide insertion corresponding to a unique BssHII restriction site to make plasmid pN2Δ. The second mutation, AUG−, was a single U21G change that eliminated synthesis of the NoV capsid protein. Plasmids pN2H and pN2G contain the yeast HIS3 and mammalian-codon optimized GFP coding regions, respectively, introduced at the unique BssHII restriction site in pN2D.
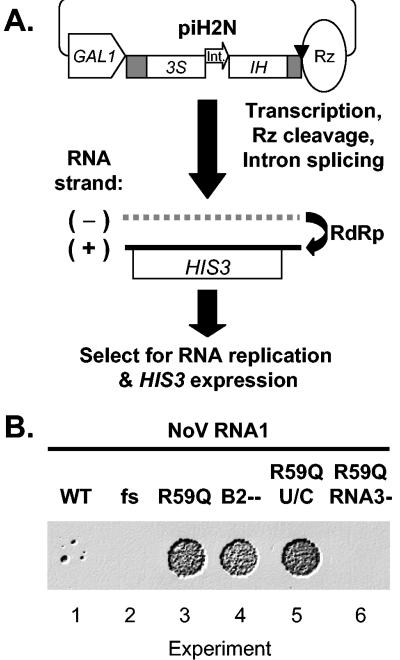
We constructed a DNA plasmid, piH2N, for RNA polII-mediated expression of negative-sense NoV RNA2 encoding the yeast HIS3 gene by reversing the orientation of the N2H cDNA relative to the GAL1 promoter. To prevent a possible cryptic positive-sense RNA transcript from serving as an mRNA for HIS3, we introduced an intron into the HIS3 coding region of pH2N to generate plasmid piH2N, as previously described for the analogous FHV plasmid, piH2F (28). Specifically, the artificial yeast intron in piH2F (cloned at the MscI site of the HIS3 ORF) was replaced with the yeast actin (ACT1) intron (11, 25) and later introduced into pH2N using the same restriction site. The general structure of the resulting plasmid piH2N is shown schematically in Fig. 4A.
FIG. 4.
Selection of yeast colonies dependent on NoV RNA replication. Panel A is a schematic. A DNA plasmid piH2N for RNA polII-mediated expression of negative-sense NoV RNA2 encoding the yeast HIS3 gene is shown at the top; symbols for sequence elements are as described for Fig. 2A. The open arrowhead labeled “Int.” indicates the yeast ACT1 intron, cloned in the antisense orientation relative to RNA2. Below, horizontal lines represent the N2H RNA with polarity indicated. The open box represents the HIS3 ORF. Panel B shows the results of selecting for HIS3-positive colonies following transient induction in galactose-containing medium. In the first four experiments, the RNA1 plasmids encoded either WT RNA1, RNA1 containing a frame-shifted polymerase (fs), an arginine-to-glutamine mutation at position 59 of the B2 ORF (R59Q), or a B2 null mutant (B2−−). In experiments 5 and 6, the RNA1 plasmids encoded the R59Q mutant in the context of a U-to-C mutation at position 1274 (U/C, experiment 5) or a 4-nt mutation that eliminates RNA3 synthesis (RNA3−, experiment 6). Yeast cells containing piH2N in addition to plasmids encoding the indicated RNA1 were induced for 2 days on galactose-containing liquid medium. An aliquot of each, containing an equal number of cells, was used to seed cultures of dextrose-containing liquid medium lacking histidine, and 2 days later, aliquots of this culture were spotted onto dextrose-containing solid medium lacking histidine. Plates were incubated at 30°C for 4 days and photographed. The image colors were digitally reversed to improve contrast.
RNA and DNA analyses.
Yeast total RNA extraction, formaldehyde denaturation, Northern blot transfer to Nytran nylon membranes (Schleicher and Schuell), and hybridization were performed as described previously (20, 24). Strand-specific 32P-labeled RNA probes for NoV were generated by in vitro transcription from PCR products containing bacteriophage RNA polymerase promoters, as described previously (32). Probes contained nt 2732 to 3204 of the appropriate sense of NoV RNA1 or nt 1 to 22 and 1100 to 1336 of the appropriate sense NoV RNA2, respectively (16). The results were visualized with a Molecular Dynamics PhosphorImager and quantitated using ImageQuant software (Amersham Biosciences Products). Isolation of genomic RNA from NoV virions was performed as described previously (16). RT-PCR was performed using a One-Step RT-PCR kit (QIAGEN) and primers specific for NoV RNA3, according to the manufacturer's instructions.
RESULTS
NoV RNA replication was initiated by transfection of virion RNA into yeast spheroplasts.
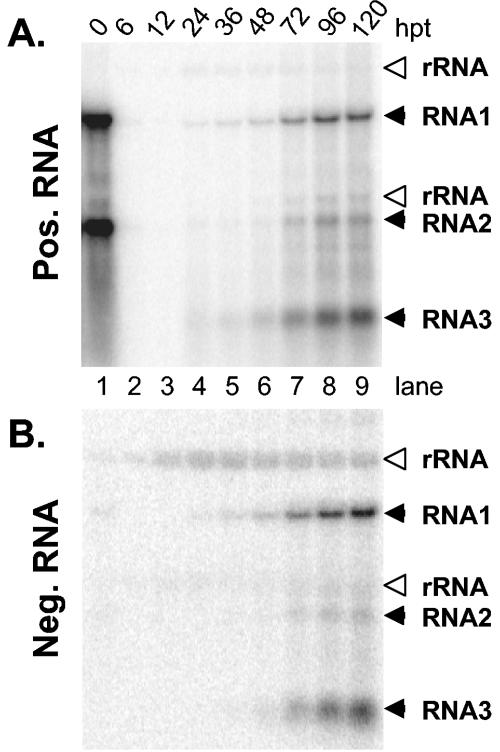
To determine if yeast cells could support the replication of NoV RNA, yeast spheroplasts were transfected with RNA isolated from NoV virions and incubated at 30°C for 5 days. At the times indicated at the top of Fig. 1A, a portion of the culture was harvested and total cellular RNAs were isolated. These RNAs were analyzed by Northern blot hybridization with probes specific for the positive and negative strands of NoV genomic RNAs and subgenomic RNA3, which is identical in sequence to the last 473 nt of NoV RNA1. Input positive-sense genomic RNA1 and RNA2 were detected immediately after transfection (Fig. 1A, lane 1). However, by 6 h posttransfection (hpt) the input material had been degraded to background levels (Fig. 1A, lane 2). Newly synthesized genomic RNAs 1 and 2 appeared by 24 hpt and increased in abundance until at least 120 hpt (Fig. 1A, lanes 4 to 9). This increase in RNAs 1 and 2 was accompanied by the appearance of subgenomic RNA3, as well as the negative-strand RNA1, RNA2, and RNA3 replication intermediates (Fig. 1B, lanes 4 to 9). The appearance of these intermediates, together with the increase in positive-sense genomic and subgenomic RNAs, indicates that NoV RNA was replicating in the yeast cells. The negative-strand-specific RNA1 and RNA2 probes weakly cross-reacted with positive-sense RNAs (Fig. 1B, lane 1). However, considering the amount of positive-sense material in each lane, the majority of the signals detected at 6 hpt and later must be due to authentic negative-sense RNA. Unexpectedly, the RNA2-to-RNA1 ratio for both positive-sense and negative-sense RNA was less than the 1:1 ratio normally observed during replication in insect cells (17) and in RNA isolated from virions (Fig. 1A, compare lanes 7 to 9 with lane 1). Instead, we observed RNA2-RNA1 ratios of approximately 1:5 and 1:2 for positive and negative strands, respectively. The same skewed ratio was observed when RNA replication was initiated from infectious cDNAs (see below), suggesting that perhaps, in yeast, replication of NoV RNA1 was preferential or that for some reason RNA2 replication was less robust.
FIG. 1.
Northern blot hybridization analysis of NoV RNA replication in yeast spheroplasts transfected with viral RNA. Yeast spheroplasts were transfected with virion-derived NoV RNA and incubated at 30°C. At various times posttransfection (hpt), total yeast RNA was extracted, separated on denaturing agarose-formaldehyde gels, and transferred to nylon membranes. Viral RNAs, indicated with solid arrowheads, were detected with strand-specific riboprobes for the 3′ end of RNA1 (corresponding to subgenomic RNA3) and for RNA2. Panel A shows positive-sense RNA. Panel B shows negative-sense RNAs. Detection of negative strands required loading of three times more RNA and imaging at fourfold higher intensity than needed to detect positive strands. Open arrowheads indicate ribosomal RNAs, which cross-reacted weakly with the probes.
Plasmid-expressed NoV RNA1 replicated to high levels in yeast and previously characterized mutations altered the ratio of RNA3 to RNA1 as expected.
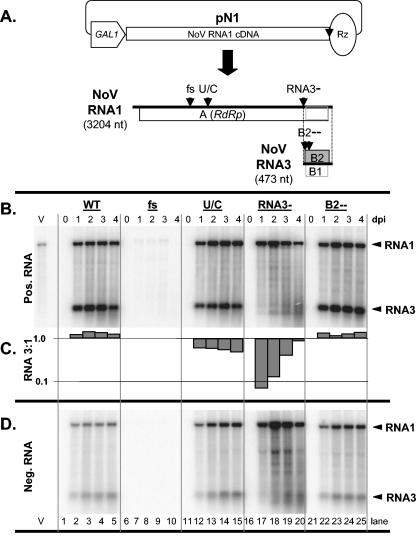
Infectious cDNA clones have been recovered for NoV genomic RNAs, RNAs 1 and 2 (16). By using the NoV RNA1 cDNA, we constructed plasmid pN1 for the in vivo transcription of NoV RNA1 from the yeast GAL1 promoter (Fig. 2A), a strong inducible polII promoter used previously for the expression of FHV RNA1 in the context of plasmid pF1 (29). Promoter positioning was based on pF1, with the 5′-most nucleotide of both viral cDNAs in the same position relative to the promoter. As in pF1, the resulting positive-sense RNA transcripts were designed to contain the self-cleaving HDV ribozyme (27), positioned to cleave at the exact 3′ terminus of NoV RNA1. This plasmid was induced in yeast and total RNA was harvested at 0, 1, 2, 3, and 4 days postinduction (dpi). Northern blot hybridization with probes specific for RNAs 1 and 3 revealed galactose-dependent bands corresponding in size to positive-sense RNA1 and subgenomic RNA3 (Fig. 2B, lanes 1 to 5) and their negative-sense counterparts (Fig. 2D, lanes 1 to 5). Since RNA3 and the negative-sense RNAs are products of RNA replication, the results of the Northern blot hybridization support the claim that plasmid-expressed NoV RNA1 replicated in yeast.
FIG. 2.
Northern blot hybridization analysis of plasmid-initiated NoV RNA1 replication in yeast cells. Panel A is a schematic showing a DNA plasmid pN1 for RNA polII-mediated transcription of NoV RNA1 and its expected DNA-dependent and RNA-dependent transcription products. An open pentagon, box, and oval represent the inducible yeast GAL1 promoter, viral RNA1 cDNA, and HDV anti-genomic ribozyme cDNA, respectively. NoV RNA1 and RNA3 are represented approximately to scale (sizes indicated) and open boxes represent their ORFs. Vertical arrowheads indicate the positions of four mutations that result in defects in accumulation of RNA1 and/or RNA3. These mutations are (i) a frameshift (fs) in the RdRp ORF caused by the insertion of 4 nt at position 982; (ii) a U/C mutation (U/C) at position 1274 that decreases RNA3 synthesis; (iii) mutation of 4 nt (2731 to 2734) that eliminates RNA3 synthesis (RNA3−); and (iv) mutation of 3 nt between positions 2745 and 2758 that closes the B2 ORF (B2−−). Panels B and D show Northern blot hybridizations for positive- and negative-sense RNAs, respectively. Total RNA was extracted from induced yeast containing the indicated NoV RNA1 plasmid at 0, 1, 2, 3, or 4 dpi and analyzed by Northern blot hybridization as described for Fig. 1, except that only the RNA1/RNA3-specific probes were used. Lane V contained genomic RNA isolated from NoV virions. Arrowheads indicate RNA1 and RNA3. Panel C shows the ratio of the positive-sense RNA3 to RNA1 for each lane in panel B, plotted on a log scale.
Quantitation revealed that NoV RNA1 replication was robust, with more than 104 copies per cell (data not shown). This level of expression is comparable to results obtained using pF1 for FHV plasmid-based expression. Indeed as determined by ethidium bromide staining of RNAs separated on nondenaturing agarose gels, the RNAs synthesized from pN1 were equally or more abundant than those derived from pF1 (data not shown). The NoV RNA1 and RNA3 bands were dependent on the virus-encoded RdRp, since a frameshift early in the polymerase ORF eliminated their synthesis (Fig. 2B and D, lanes 6 to 10). Instead, a positive-sense RNA species slightly larger than RNA1 was detected (Fig. 2B, lanes 7 to 9), which likely corresponds to primary transcripts containing the uncleaved HDV ribozyme (8).
Two mutations in NoV RNA1 have been shown previously to alter the level of RNA3 relative to that of RNA1 in mammalian cells. The point mutation U1274C reduces RNA3 levels to 20% of WT levels (16) and a quadruple mutation that changes four consecutive nucleotides flanking the RNA3 5′ end (RNA3−) reduces positive RNA3 synthesis to undetectable levels (17). Incorporation of U1274C or RNA3− into pN1 resulted in phenotypes in yeast that were similar to those observed in mammalian cells (Fig. 2B, C, and D; compare lanes 1 to 5 with lanes 11 to 15 and 16 to 20, respectively). Quantitation revealed that the RNA3-to-RNA1 ratio decreased by greater than 12-fold for RNA3− (Fig. 2C, lanes 16 to 20). In the hamster BSR T7/5 cell system, this same mutation caused the RNA3-to-RNA1 ratio to decrease by greater than 97% (17). The absolute RNA1 levels varied to a greater degree for RNA3− than for the other RNA1 clones, peaking at 2 dpi and decreasing thereafter (Figs. 2B and D, lanes 18 to 20).
For FHV, the RNA3-encoded B2 protein has been shown to suppress RNAi and is required for FHV RNA replication in Drosophila cells (21). In contrast, FHV B2 is not required for replication in S. cerevisiae (29). This difference in the need for FHV B2 is thought to be due to the fact that S. cerevisiae lacks the conserved genes that encode the RNAi pathway (3). Accordingly, a triple mutant, NoV B2−−, that was unable to express either form of the NoV B2 protein (16, 17) replicated to levels 1.5 times higher than WT NoV RNA1 and displayed an RNA3-to-RNA1 ratio similar to that of WT NoV RNA1 (Fig. 2A, B, and C, compare lanes 1 to 5 to lanes 21 to 25).
Plasmid-expressed NoV RNA2 replicated in yeast.
By using the NoV RNA2 cDNA, we constructed plasmid pN2 for the in vivo transcription of positive-sense NoV RNA2 from the yeast GAL1 promoter (Fig. 3A). The promoter and ribozyme positioning of this plasmid were identical to those of plasmid pF2U (29), a plasmid that directs the in vivo expression of an FHV RNA2 replicon containing the yeast URA3 gene. Yeast cells transformed with pN1 and pN2 were induced with galactose as described for Fig. 2. Total RNA was harvested at 0 and 1 dpi and subjected to analysis by Northern blot hybridization as before, except that probes specific for RNA2 were used (Fig. 3B). Galactose-dependent RNAs corresponding in size to positive- (Fig. 3B, lane 2) and negative-sense RNA2 (data not shown) were detected at 1 dpi. The abundance of NoV RNA2 relative to that of NoV RNA1 in these experiments, where RNA replication was initiated from plasmids, was similar to that observed following transfection of virion RNA into yeast spheroplasts (Fig. 1; also data not shown). Furthermore, a similar ratio of RNA2 to RNA1 was maintained over a time course of at least 4 days (data not shown). A slightly larger positive-sense RNA was also observed (Fig. 3B) and likely corresponded to transcripts containing the uncleaved HDV ribozyme (8). This idea was supported by the observation that this RNA species hybridized to an HDV-specific probe and decreased in abundance with time postinduction (data not shown).
FIG. 3.
Northern blot hybridization analysis of plasmid-initiated NoV RNA2 replication in yeast cells. Panel A is a schematic of a DNA plasmid pN2 for RNA polII-mediated transcription of NoV RNA2 and its expected DNA-dependent and RNA-dependent transcription products; symbols for sequence elements are as described for Fig. 2A. The open box in RNA2 represents the capsid ORF. The relative positions of several RNA2 derivatives are indicated: a mutation of the translational start codon (AUG−), a derivative in which nt 53 to 476 were deleted (N2Δ), an N2Δ deletion derivative encoding the yeast HIS3 ORF (N2H), and an N2Δ deletion derivative encoding mammalian codon-optimized GFP (N2G). Panel B shows a Northern blot of positive-sense RNAs. Total RNA was extracted from induced yeast containing a plasmid encoding WT NoV RNA1 and the indicated RNA2 plasmid at 0 and 1 dpi and analyzed by Northern blot hybridization as described for Fig. 1, except that only riboprobes specific for positive-sense RNA2 were used. For each construct, the position of the full-length RNA is indicated with a solid arrowhead at the right, while the open arrowhead indicates the primary transcript before cleavage by the HDV ribozyme (see Materials and Methods).
Characterization of RNA2 deletion derivatives as replicons for expression of heterologous sequences.
Defective-interfering RNAs arise spontaneously during passage of nodaviruses and their RNAs at high multiplicity (36). Naturally occurring derivatives of NoV RNA2 with internal deletions have previously been identified in our laboratory by RT-PCR analysis (T. Gratsch and L. A. Ball, unpublished observations). Independent NoV RNA2 clones contained deletions downstream of nt 52 or upstream of nt 422, suggesting that nt 53 to 423 might be dispensable for RNA2 replication. These nucleotides were deleted from pN2 and replaced by a six-nucleotide insertion corresponding to a unique BssHII restriction site to make plasmid pN2Δ, which directed synthesis of the RNA N2Δ (Fig. 3A). Northern blot hybridization revealed galactose-dependent bands corresponding in size to positive (Fig. 3B, compare lanes 3 and 4) and negative (data not shown) strands of N2Δ. Surprisingly, N2Δ was approximately fivefold more abundant than WT NoV RNA2 isolated from pN2-induced yeast (Fig. 3B, compare lanes 4 and 2, respectively). This RNA also differs from WT RNA2 in size and coding capacity: N2Δ RNA is 364 nt smaller than WT RNA2. The first ORF of N2Δ contained only 11 N-terminal codons of the NoV capsid protein and two novel codons (encoding arginine and alanine, respectively) from the six-nucleotide insertion, followed by a stop codon. An even smaller NoV RNA2 derivative (N2ΔΔ) that lacked nt 53 to 1066 and encoded an ORF of only 50 codons accumulated to similar levels (data not shown).
Analysis of N2Δ suggested that the presence of the NoV RNA2 ORF inhibited RNA2 replication. To test this hypothesis, we constructed a plasmid for the expression of an RNA2 derivative with a single U-to-G substitution at position 21. The encoded AUG− RNA lacks the authentic capsid ORF start codon. Northern blot hybridization (Fig. 3B, lanes 5 and 6) revealed that AUG− behaved similarly to N2Δ and replicated approximately sixfold more abundantly than WT NoV RNA2 (Fig. 3B, compare lane 6 with lane 2). RNA1 replication was unaffected in these experiments.
FHV RNA2-based replicons have been used previously to express yeast nutritional markers such as URA3 and HIS3 (28, 30). Replication of these heterologous mRNAs by RdRp expressed by FHV RNA1 allows RNA replication to be tied to yeast phenotypes. To accomplish the same for NoV, we inserted the yeast HIS3 gene into the N2Δ vector. The resulting RNA, N2H, encodes the first 11 amino acids of NoV capsid protein and two novel amino acids (arginine and valine) fused to HIS3. Similar fusion proteins have been previously expressed from FHV RNA2-based vectors (28, 30). Northern blot hybridization revealed N2H-sized RNA species only after induction with galactose (Fig. 3B, lanes 7 and 8). These RNAs accumulated to lower levels than N2Δ, perhaps due to the insertion of the heterologous HIS3 sequences. Similar levels of accumulation were observed after induction for N2G (Fig. 3B, lanes 9 and 10), an N2Δ derivative encoding a similar NoV capsid-GFP fusion protein. The encoded HIS3 and GFP fusion proteins were functional (data not shown), as shown previously for the FHV RNA2-derived fusions (28, 30).
The NoV B2 ORF inhibited colony formation.
We previously described a system for the in vivo transcription of HIS3-encoding FHV RNA2 replicons from cDNAs (28). These replicons were transcribed in the negative-sense from the inducible GAL1 promoter by yeast RNA polII. To ensure that only negative-sense primary transcripts could serve as templates for replication-mediated HIS3 expression, a yeast intron was inserted into the HIS3 ORF in such a way that it could be spliced only from negative-sense transcripts. The FHV RdRp copied the spliced polII RNA transcript into messenger-sense RNA and amplified it. This RNA complemented a deletion of the chromosomal copy of HIS3 and yielded RdRp-dependent yeast colonies (28).
The analogous system was constructed for polII transcription of negative-sense N2H, the NoV RNA2 HIS3 replicon, except that the yeast actin (ACT1) intron (11, 25) was used to construct plasmid piH2N (shown schematically in Fig. 4A), as described in Materials and Methods. Viral RNAs were expressed in vivo by transient induction in galactose-containing medium as before. RNA replication-dependent colonies were then selected on dextrose-containing medium lacking histidine. Although HIS+ colonies were detected, the NoV system was approximately 104-fold less efficient than the corresponding FHV system and yielded very few colonies (Fig. 4B, experiment 1). However, colony formation was RNA replication-dependent because it was eliminated by a frameshift mutation in the NoV RdRp coding region (Fig. 4B, experiment 2).
The low frequency of colony formation in the NoV system suggested the possibility that colonies might arise only after mutation. To identify possible mutations in RNA1 or RNA3, an RNA3 cDNA was prepared by RT-PCR amplification using as a template total RNA isolated from one of the rare HIS+ colonies. The results of DNA sequencing revealed a single G-to-A change at nt 188 in RNA3, which corresponds to position 2919 in RNA1. In the B2 ORF this mutation replaced arginine 59 with glutamine, whereas it was translationally silent in the overlapping polymerase ORF. When the mutation was incorporated into an otherwise WT pN1 background, it supported colony formation at a high frequency (Fig. 4B, experiment 3) similar to that previously described for WT pF1 and piH2F in the corresponding FHV system (28).
These results suggested that protein B2 (and arginine 59) or RNA3 (and nucleotide 188) inhibited NoV RNA replication-dependent colony formation. To determine if the mutation was working at the protein or the RNA level, a previously described NoV1 mutant that eliminates B2 expression, B2−− (16, 17), was tested for its ability to support colony formation. When the B2−− mutations were incorporated into the pN1 expression plasmid, the resulting RNA1 mutant supported colony formation at a high frequency (Fig. 4B, experiment 4). These results contrast with the behavior of the FHV system, which did not select for mutations in the B2 ORF (data not shown). Furthermore, an FHV B2−− mutant was similar to WT FHV RNA1 at supporting the formation of RNA replication-dependent colonies (data not shown).
For FHV, the synthesis of RNA3 is essential for RNA2 replication (9). An FHV RNA1 mutant unable to produce RNA3 will neither replicate the FHV RNA2 derivative that carries the HIS3 ORF nor support yeast colony formation (data not shown). To examine the importance of NoV RNA3 synthesis for NoV RNA2 replication, we tested the ability of the RNA1 mutants U1274C and RNA3− to support replication of the NoV RNA2-derivative iH2N and to form colonies. To eliminate possible detrimental effects of the B2 ORF, the R59Q mutation was incorporated into plasmids pN1U/C and pN1RNA3−. In the R59Q background, the U/C mutant, but not RNA3−, was found to support colony formation (Fig. 4B, experiments 5 and 6), suggesting that NoV RNA3 was required for RNA2 replication. This observation is consistent with previous results obtained in mammalian cells, in which coexpression of RNA2 with RNA1 mutants U1274C and RNA3− resulted in RNA2 replication levels that were reduced relative to those seen with WT RNA1 (16).
NoV RNA1 supported weak replication of FHV RNA2, but not vice versa.
Infectious reassortant viruses for NoV and FHV have not been recovered previously, despite extensive efforts (10). These observations could be due to an inability either of the RdRps to cross-replicate the heterologous RNA or of the capsid proteins to encapsidate the heterologous RNA. Yeast colony formation was used to examine the ability of each RdRp to replicate the homologous and heterologous RNA2-derived replicons (Fig. 5). As previously described (28), FHV RNA1 supported homologous iH2F-dependent colony formation (Fig. 5, upper left panel). However, FHV RNA1 was unable to support heterologous iH2N-dependent colony formation (Fig. 5, lower left panel). NoV RNA1 bearing the R59Q mutation in B2 also supported homologous iH2N-dependent colony formation (Fig. 5, lower right panel), as seen in Fig. 4. Unexpectedly, the heterologous combination of NoV RNA1 bearing the R59Q mutation and iH2F yielded occasional colonies (Fig. 5, upper right panel). These NoV RNA1/iH2F-dependent colonies arose at a frequency approximately 104-fold lower than the corresponding FHV RNA1/iH2F colonies (Fig. 5, upper left panel). The similarity of this frequency with the observed rate for colony formation again suggests selection for mutations.
FIG. 5.
Selection of yeast colonies dependent on RNA2 replication by FHV or NoV RdRp. Yeast cells were transiently induced and plated as described for Fig. 4B. The identity of each RNA1 species is indicated at the top. Note that NoV RNA1 encoded the R59Q mutation in the B2 coding region, whereas FHV RNA1 was WT with respect to B2. The identity of each RNA2 species is indicated at the left.
DISCUSSION
Here we have demonstrated the ability of NoV to replicate and transcribe its entire genome in yeast. To date, FHV and NoV are the only two positive-sense RNA viruses to replicate their entire genomes in yeast, although several other viruses (with RNA or DNA genomes) can undergo partial replication in yeast cells (2, 15, 26, 31, 35). NoV is the only virus infectious to mammals able to fully replicate its genome in yeast. NoV RNA1 replication was as robust as that of FHV RNA1. NoV RNA1 replicated to approximately 104 copies per cell, making the viral RNAs easily observed by staining with ethidium bromide. Three previously described NoV RNA1 mutations exhibited altered RNA3-to-RNA1 ratios in yeast, as expected from results obtained in mammalian and insect cells (16, 17).
In contrast with FHV, whose genomic RNAs accumulated to equimolar levels in all cell types examined, the ratio of NoV RNA1 to RNA2 was found to vary in yeast. Accumulation of the NoV genomic segments was not equimolar, either in yeast transfected with genomic RNAs or in yeast induced to transcribe RNAs from infectious cDNAs. Similarly, NoV RNA1 replicated to levels that were somewhat higher than that of RNA2 in transfected BHK21 cells (16) and to even higher levels in several insect cell lines. However, the NoV genomic segments accumulated to equimolar levels in mosquito TRA-171 cells (K. L. Johnson, B. D. Price, and L. A. Ball, unpublished observations). Together, these results suggest that the relative ratio of the NoV genomic segments produced during RNA replication varies with cell type and/or experimental conditions, whereas similar behavior was not observed for FHV RNAs.
In yeast, removal of the RNA2-encoded capsid ORF greatly increased the accumulation of RNA2. This result suggested that RNA2 translation might inhibit RNA replication in yeast. It is possible that ribosomes may stall on the RNA2 mRNA, perhaps due to inappropriate codon usage (19), and consequently RNA2 may be sequestered from the RNA replication machinery. Alternatively, the capsid protein precursor may somehow interfere with NoV RNA replication in yeast.
Since NoV RNAs replicated in yeast, we established a HIS3-based system in which colony formation depended on RNA replication. Unexpectedly, the WT NoV B2 protein was found to inhibit colony formation. An R59Q (G2919A) mutation in the NoV B2 ORF was the first mutation identified that relieved this inhibition, but many different additional B2 mutants have since been identified (B.D. Price and L. A. Ball, unpublished data).
NoV B2 inhibited RNA replication-dependent colony formation, whereas FHV B2 does not in the analogous FHV system. Although NoV B2 (137 and 134 amino acids) and FHV B2 (106 amino acids) share only 29% sequence identity (17), it is likely that these two proteins share common functions. Certainly, both have been shown to inhibit RNAi in some other host cells (17, 21, 22). Other viral inhibitors of RNAi have been shown to bind double-stranded RNA (23) and the nodavirus B2 protein may also act by this mechanism, thus sequestering double-stranded RNA from the RNAi machinery. However, the differences between FHV and NoV observed in our system suggests that the B2 proteins encoded by the two viruses may have additional functions or vary in their efficiencies to enact a common function. In addition, these proteins may be expressed at different levels in yeast, perhaps due to different codon usage or efficiency of translational initiation. Indeed, following RNA1 induction, NoV B2, but not FHV B2, was easily detected in total yeast extracts by Coomassie blue staining of sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels (data not shown). The NoV B2 protein itself is not toxic to the yeast. Yeast cells carrying WT pN1 alone readily grow, and form colonies, on galactose-containing medium (data not shown).
We explored the RNA3 dependence of NoV RNA2 replication by using the yeast system. A RNA1 mutant (U1274C) that reduces RNA2 replication to 20% in mammalian cells (16) was found to support N2H-dependent colony formation with similar efficiency as WT RNA1. However, an NoV RNA1 mutant (RNA3−) unable to transcribe NoV RNA3 failed to support RNA replication-dependent colony formation. This suggests that NoV RNA3 is required for RNA2 replication, as suggested previously for NoV RNAs in mammalian cells (16). This requirement for RNA3 has also been observed for FHV in a plasmid-based RNA replication system in mammalian cells (9) and in a similar yeast system (data not shown). The NoV RNA3− mutant contained four nucleotide changes, resulting in two missense mutations (R904K and V905I) in the overlapping carboxy-terminal region of the RdRp (17). These amino acid changes apparently did not to alter polymerase function, either in yeast (Fig. 2B and 2D, lanes 16 to 20) or in mammalian cells (17). Instead, these mutations likely disrupt cis-acting RNA signals necessary for RNA3 transcription.
We tested the ability of NoV RNA1 and FHV RNA1 to support the replication of the heterologous RNA2 using the yeast genetic system. FHV RNA1 could not support heterologous iH2N-dependent colony formation, but NoV RNA1 bearing the R59Q mutation could support iH2F-dependent colony formation. Evidently NoV RNA3 and RdRp, respectively, could transactivate and replicate the heterologous replicon well enough to permit HIS3-dependent colony formation. However, colonies arose with low frequency, suggesting that mutation of one or the other of the viral RNAs may have been required to allow colony formation. We anticipate that identification of such mutations may shed light on the determinants of RNA template specificity between NoV and FHV.
These results clearly demonstrate the power of the yeast system for genetic identification of factors that promote or inhibit nodavirus RNA replication. We are now using this system to examine the function of the NoV B2 protein and to identify cis-acting RNA elements that allow the replication of heterologous genomic segments. In addition, we can use yeast to produce large quantities of viral and heterologous RNAs and proteins from the RNA2-based replicons. Such products could include infectious nodaviruses, heterologous virus capsid proteins and virus-like particles, and vaccine material. The ability to make such products in yeast will facilitate many avenues in basic and applied research.
Acknowledgments
We thank Melissa Kranz for excellent technical assistance and the UAB Center for AIDS Research DNA sequencing core facility. We thank the other members of our laboratory and Cindy Luongo (UAB) for many helpful discussions and critical review of the manuscript. We also thank Kim M. Keeling (UAB) and Tom Oomens (UAB) for critical review of the manuscript, and Edward C. Walthall (UAB Cystic Fibrosis Research Center) for assistance with computer graphics.
This work was supported by NIH grant R21 AI52736 to B.D.P., Cystic Fibrosis Foundation grant CFF JOHNSO03I0 to K.L.J., and NIH grant RO1 AI18270 to L.A.B.
REFERENCES
- 1.Ammerer, G. 1983. Expression of genes in yeast using the ADCI promoter. Methods Enzymol. 101:192-201. [DOI] [PubMed] [Google Scholar]
- 2.Angeletti, P. C., K. Kim, F. J. Fernandes, and P. F. Lambert. 2002. Stable replication of papillomavirus genomes in Saccharomyces cerevisiae. J. Virol. 76:3350-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aravind, L., H. Watanabe, D. J. Lipman, and E. V. Koonin. 2000. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc. Natl. Acad. Sci. USA 97:11319-11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball, L. A., J. M. Amann, and B. K. Garrett. 1992. Replication of nodamura virus after transfection of viral RNA into mammalian cells in culture. J. Virol. 66:2326-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ball, L. A., and K. L. Johnson. 1999. Reverse genetics of nodaviruses. Adv. Virus Res. 53:229-244. [DOI] [PubMed] [Google Scholar]
- 6.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 7.Dasmahapatra, B., R. Dasgupta, K. Saunders, B. Selling, T. Gallagher, and P. Kaesberg. 1986. Infectious RNA derived from transcription from cloned cDNA copies of the genomic RNA of an insect virus. Proc. Natl. Acad. Sci. USA 83:63-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckerle, L. D., C. G. Albarino, and L. A. Ball. 2003. Flock House virus subgenomic RNA3 is replicated and its replication correlates with transactivation of RNA2. Virology 317:95-108. [DOI] [PubMed] [Google Scholar]
- 9.Eckerle, L. D., and L. A. Ball. 2002. Replication of the RNA segments of a bipartite viral genome is coordinated by a transactivating subgenomic RNA. Virology 296:165-176. [DOI] [PubMed] [Google Scholar]
- 10.Gallagher, T. M. 1987. Synthesis and assembly of nodaviruses. Ph.D. thesis. University of Wisconsin—Madison, Madison.
- 11.Gallwitz, D., and I. Sures. 1980. Structure of a split yeast gene: complete nucleotide sequence of the actin gene in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 77:2546-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garzon, S., and G. Charpentier. 1991. Nodaviridae, p. 351-370. In J. R. Adams and J. R. Bonami (ed.), Atlas of invertebrate viruses. CRC Press, Boca Raton, Fla.
- 13.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 14.Hill, J. E., A. M. Myers, T. J. Koerner, and A. Tzagoloff. 1986. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2:163-167. [DOI] [PubMed] [Google Scholar]
- 15.Janda, M., and P. Ahlquist. 1993. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell 72:961-970. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, K. L., B. D. Price, and L. A. Ball. 2003. Recovery of infectivity from cDNA clones of Nodamura virus and identification of small nonstructural proteins. Virology 305:436-451. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, K. L., B. D. Price, L. D. Eckerle, and L. A. Ball. 2004. Nodamura virus nonstructural protein B2 can enhance viral RNA accumulation in both mammalian and insect cells. J. Virol. 78:6698-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, K. N., K. L. Johnson, R. Dasgupta, T. Gratsch, and L. A. Ball. 2001. Comparisons among the larger genome segments of six nodaviruses and their encoded RNA replicases. J. Gen. Virol. 82:1855-1866. [DOI] [PubMed] [Google Scholar]
- 19.Kozak, M. 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234:187-208. [DOI] [PubMed] [Google Scholar]
- 20.Leeds, P., S. W. Peltz, A. Jacobson, and M. R. Culbertson. 1991. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 5:2303-2314. [DOI] [PubMed] [Google Scholar]
- 21.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 22.Li, W.-X., H. Li, R. Lu, F. Li, M. Dus, P. Atkinson, E. W. A. Brydon, K. L. Johnson, A. Garcia-Sastre, L. A. Ball, P. Palese, and S.-W. Ding. 2004. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. USA 101:1350-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lichner, Z., D. Silhavy, and J. Burgyan. 2003. Double-stranded RNA-binding proteins could suppress RNA interference-mediated antiviral defences. J. Gen. Virol. 84:975-980. [DOI] [PubMed] [Google Scholar]
- 24.Newman, T. C., M. Ohme-Takagi, C. B. Taylor, and P. J. Green. 1993. DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell 5:701-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng, R., and J. Abelson. 1980. Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 77:3912-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panavas, T., and P. D. Nagy. 2003. Yeast as a model host to study replication and recombination of defective interfering RNA of Tomato bushy stunt virus. Virology 314:315-325. [DOI] [PubMed] [Google Scholar]
- 27.Perrotta, A. T., and M. D. Been. 1991. A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature (London) 350:434-436. [DOI] [PubMed] [Google Scholar]
- 28.Price, B. D., P. Ahlquist, and L. A. Ball. 2002. DNA-directed expression of an animal virus RNA for replication-dependent colony formation in Saccharomyces cerevisiae. J. Virol. 76:1610-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price, B. D., M. Roeder, and P. Ahlquist. 2000. DNA-directed expression of functional Flock House virus RNA1 derivatives in Saccharomyces cerevisiae, heterologous gene expression, and selective effects on subgenomic mRNA synthesis. J. Virol. 74:11724-11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price, B. D., R. R. Rueckert, and P. Ahlquist. 1996. Complete replication of an animal virus and maintenance of expression vectors derived from it in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:9465-9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raghavan, V., P. S. Malik, N. R. Choudhury, and S. K. Mukherjee. 2004. The DNA-A component of a plant geminivirus (Indian mung bean yellow mosaic virus) replicates in budding yeast cells. J. Virol. 78:2405-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Scherer, W. F., and H. S. Hurlbut. 1967. Nodamura virus from Japan: a new and unusual arbovirus resistant to diethyl ether and chloroform. Am. J. Epidemiol. 86:271-285. [DOI] [PubMed] [Google Scholar]
- 34.Selling, B. H., R. F. Allison, and P. Kaesberg. 1990. Genomic RNA of an insect virus directs synthesis of infectious virions in plants. Proc. Natl. Acad. Sci. USA 87:434-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao, K. N., and I. H. Frazer. 2002. Replication of bovine papillomavirus type 1 (BPV-1) DNA in Saccharomyces cerevisiae following infection with BPV-1 virions. J. Virol. 76:3359-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhong, W. 1993. Flock House virus, a small insect ribovirus: replication and encapsidation of RNA2. Ph.D. thesis. University of Wisconsin—Madison, Madison.