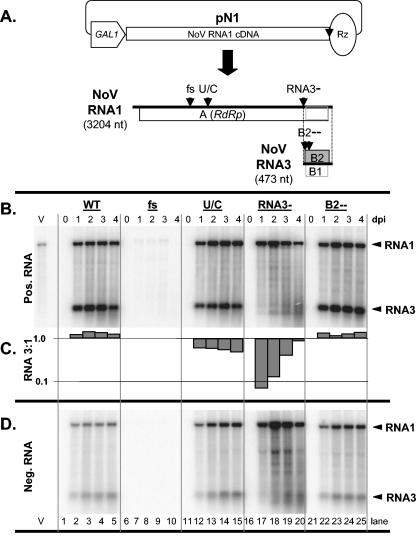
FIG. 2.
Northern blot hybridization analysis of plasmid-initiated NoV RNA1 replication in yeast cells. Panel A is a schematic showing a DNA plasmid pN1 for RNA polII-mediated transcription of NoV RNA1 and its expected DNA-dependent and RNA-dependent transcription products. An open pentagon, box, and oval represent the inducible yeast GAL1 promoter, viral RNA1 cDNA, and HDV anti-genomic ribozyme cDNA, respectively. NoV RNA1 and RNA3 are represented approximately to scale (sizes indicated) and open boxes represent their ORFs. Vertical arrowheads indicate the positions of four mutations that result in defects in accumulation of RNA1 and/or RNA3. These mutations are (i) a frameshift (fs) in the RdRp ORF caused by the insertion of 4 nt at position 982; (ii) a U/C mutation (U/C) at position 1274 that decreases RNA3 synthesis; (iii) mutation of 4 nt (2731 to 2734) that eliminates RNA3 synthesis (RNA3−); and (iv) mutation of 3 nt between positions 2745 and 2758 that closes the B2 ORF (B2−−). Panels B and D show Northern blot hybridizations for positive- and negative-sense RNAs, respectively. Total RNA was extracted from induced yeast containing the indicated NoV RNA1 plasmid at 0, 1, 2, 3, or 4 dpi and analyzed by Northern blot hybridization as described for Fig. 1, except that only the RNA1/RNA3-specific probes were used. Lane V contained genomic RNA isolated from NoV virions. Arrowheads indicate RNA1 and RNA3. Panel C shows the ratio of the positive-sense RNA3 to RNA1 for each lane in panel B, plotted on a log scale.