Figure 6.
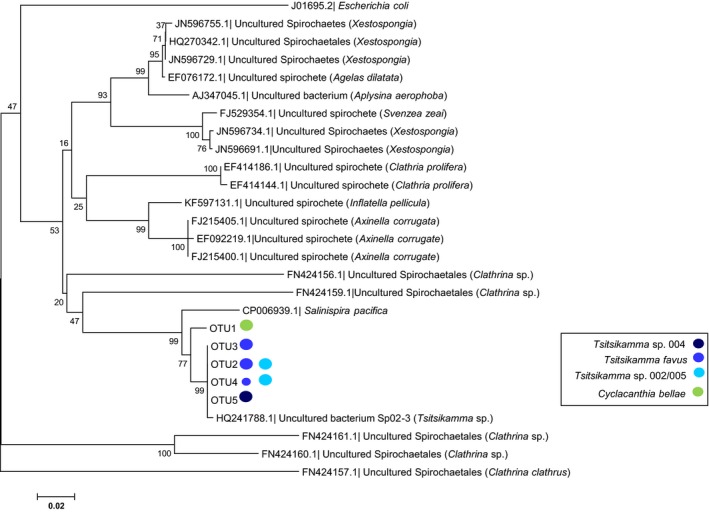
Phylogenetic relationship between dominant latrunculid Spirochaetae OTUs and other predominantly sponge‐associated Spirochaetes. Related Spirochaetae sequences were obtained from GenBank (corresponding accession number indicated) and, where possible, the sponge host is provided. The phylogenetic analysis was generated using MEGA6 software (Tamura et al., 2013) with the Neighbor‐joining method (Saitou & Nei, 1978). The 16S rRNA gene sequence of Escherichia coli (J01695) was used to root the tree. Bootstrap values, calculated based on 1,000 replicates, are indicated next to the branches (Felsenstein, 1985). Evolutionary distance was calculated using the maximum composite likelihood method (Tamura et al., 2004) based on the number of nucleotide substitutions per site over a total of 259 positions. The branch lengths correlate with evolutionary distance used to infer the phylogenetic tree. The host of each OTU is indicated by a color‐coded circle with large versus small circles indicating relative dominance
