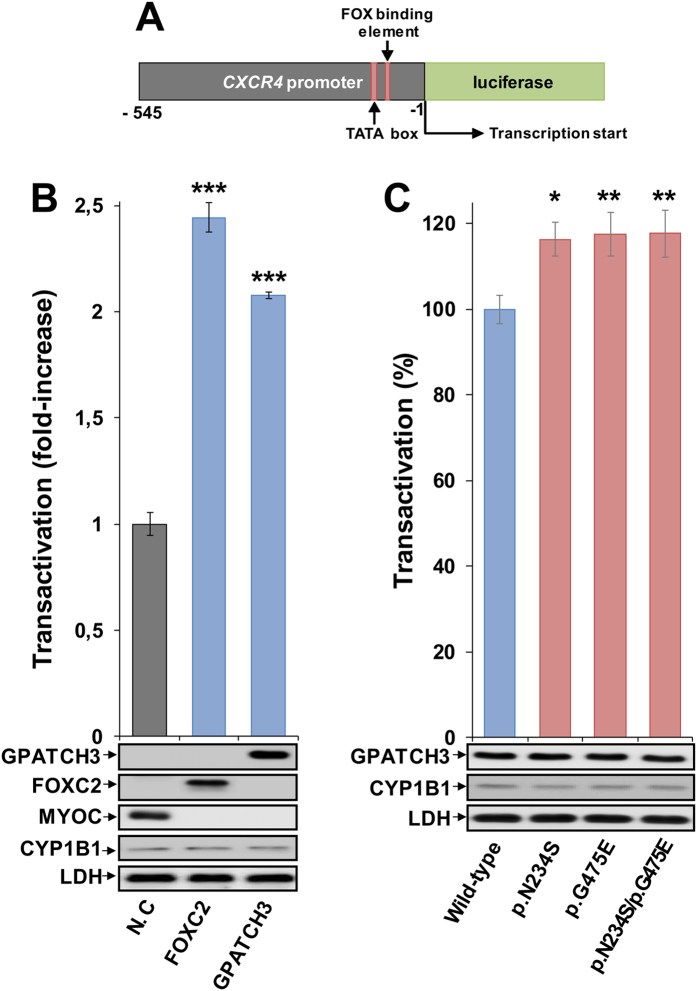
Figure 4. Transactivation of the CXCR4 promoter by GPATCH3 and increased activity of variants p.Asn234Ser and p.Gly475Glu.
(A) Scheme of the cDNA construct containing a FOX-binding element and the TATA box sequence present in the CXCR4 promoter fused to the luciferase coding region. The cDNA was cloned into the PGL3 basic vector. This construct was used as a reporter of transcriptional activity in co-transfection assays with cDNAs encoding the transcription factor FOXC2 (positive control), the extracellular protein myocilin (MYOC, negative control), the different GPATCH3 variants and the intracellular protein CYP1B1 (transfection efficiency control). The numbers below the scheme correspond to nucleotide positions. (B) cDNA constructs encoding MYOC, FOXC2 and GPATCH3 were transiently co-expressed with the reporter luciferase cDNA in HEK-293T cells. The transcriptional activity, expressed as fold increase of the luciferase activity of MYOC (negative control, N.C), was measured as indicated in the Methods section. (C) cDNA constructs encoding the different variants of GPATCH3 were transiently co-expressed with the reporter luciferase cDNA in HEK-293T cells. The transcriptional activity, expressed percentage of the luciferase activity of wild-type GPATCH3, was measured as indicated in the Methods section. The protein levels present in HEK-293T cells 24 h after transfection were determined by western immunoblot using a monoclonal anti-myc antibody. Each lane contained 15 μg of total protein obtained from the cell lysates. Transfection efficiency (CYP1B1) was assessed via western immunoblot using a monoclonal anti-myc antibody. The sample loading control, endogenous LDH, was also detected via immunoblot using an anti-LDH antibody. Values are expressed as mean ± SEM of at least three independent experiments carried out in triplicate. Asterisks indicate statistical significance as compared to the control: p < 0.02 (*); p < 0.006 (**); p < 0.001 (***). Statistical significance was calculated by Student’s t-test.