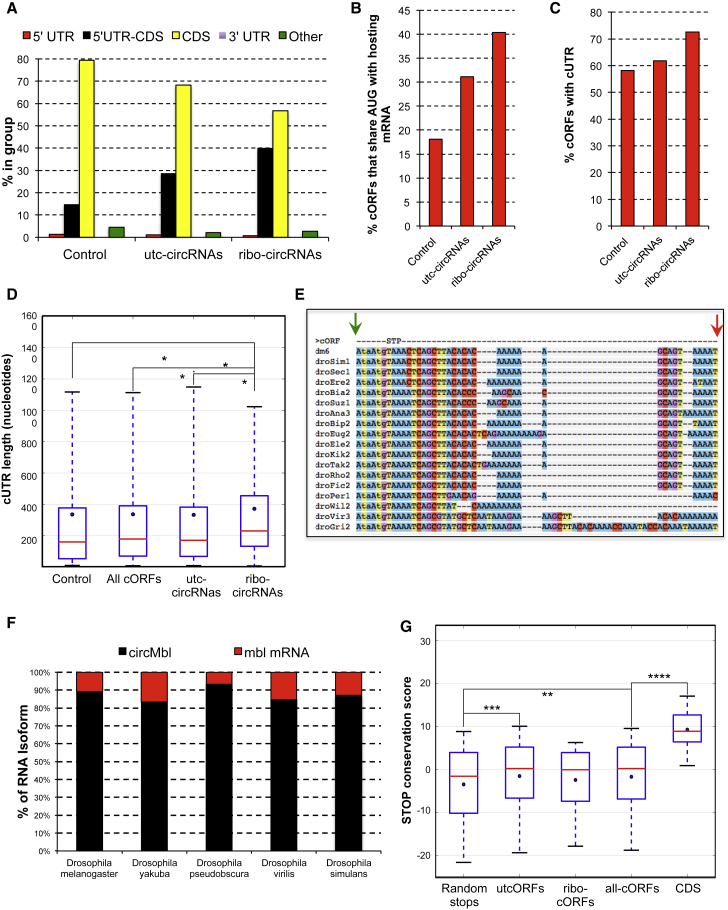
Figure 5.
Features of Fly Ribo-CircRNAs
(A) Ribo-circRNAs are strongly enriched for 5′ UTR and CDS overlap (p < 0.0055, Fisher’s exact test).
(B) Percentage of control, utc-circRNAs, and ribo-circRNAs ORFs, which share their putative start codon with their hosting linear mRNA. The differences between ribo-circRNAs and control or utc-circRNAs are statistically significant (p < 2 × 10−9 and p < 0.05, respectively, Fisher’s exact test).
(C) Percentage of control, cORFs, and ribo-circRNAs that contain cUTR. The differences between ribo-circRNAs and control or utc-circRNAs are significant (p < 0.002 and p < 0.05, respectively, Fisher’s exact test).
(D) Average length of cUTR for control groups as well as for ribo-circRNAs (p < 0.019, double-sided Mann-Whitney U test).
(E) Excerpt of a multiple species alignment of circMbl. First codon position is in uppercase, and other codon positions are in lowercase. STP, stop codon. Arrows indicate the beginning (green) and end (red) of the exon.
(F) Ratio between the linear and circular forms of the second exon of mbl in different Drosophila species. n = 2.
(G) Boxplot showing the distribution of stop codon conservation scores for circRNAs composed of CDS and 5′ UTR exonic sequences. Negative controls are randomly selected stop codons inside the same 5′ UTR stretches. CDS denotes annotated stop codons from mRNAs sharing the circRNA exons. Red lines indicate the median, and blue dots indicate the mean. Boxes show interquartile range. Whiskers show 5th to 95th percentile range. Significantly different medians are indicated by asterisks: ∗∗∗∗p < 0.0001, two-sided Mann-Whitney U test.
∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. See also Figures S5 and S6 and Tables S3, S4, S5, and S6.