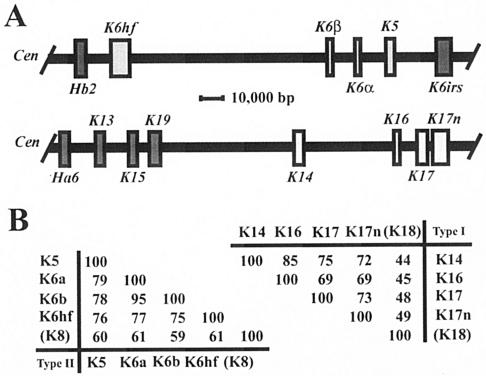
FIG. 4.
Clustering of related keratin genes on mouse chromosomes 15 and 11. (A) Organization of subsets of type II and type I keratin genes on mouse chromosomes 15 (top) and 11 (bottom), respectively. In both instances the centromere (Cen) is located on the left side. All keratin genes located in these two clusters, including the ones shown here, display the same transcriptional orientation towards the centromere. All type II keratin genes exhibit nine introns, whereas all type I genes exhibit eight introns, located in each case at identical positions within the coding sequence (data not shown; see reference 16). The genes of interest to this study are displayed in light-shaded boxes. This information was adapted from Wang et al. (48) and Tong and Coulombe (46). (B) Pairwise comparisons of the primary structures for type II (left) and type I (right) keratins expressed in the nail bed epithelium. Percent identity scores, as determined by the maximum matching routine in DNAsis version 3.5 software (Hitachi, Tokyo, Japan), are shown for the sequences encoding the nonhelical head and central rod domain of these keratins. K8 and K18, which represent the major keratin pair expressed in simple epithelia, have been included for reference purposes. The physical proximity, identical substructures, identical orientations of transcription, and obvious sequence relatedness of the type II keratin genes K5, K6α, K6β, and K6hf as well as the type I genes K14, K16, K17, and K17n strongly suggest that they were each generated through successive duplications from a common ancestral gene. The information needed to construct this figure is derived from the mouse genome browser available at http://www.ensembl.org/. The same principles apply to the human orthologs for these genes (4).